Abstract
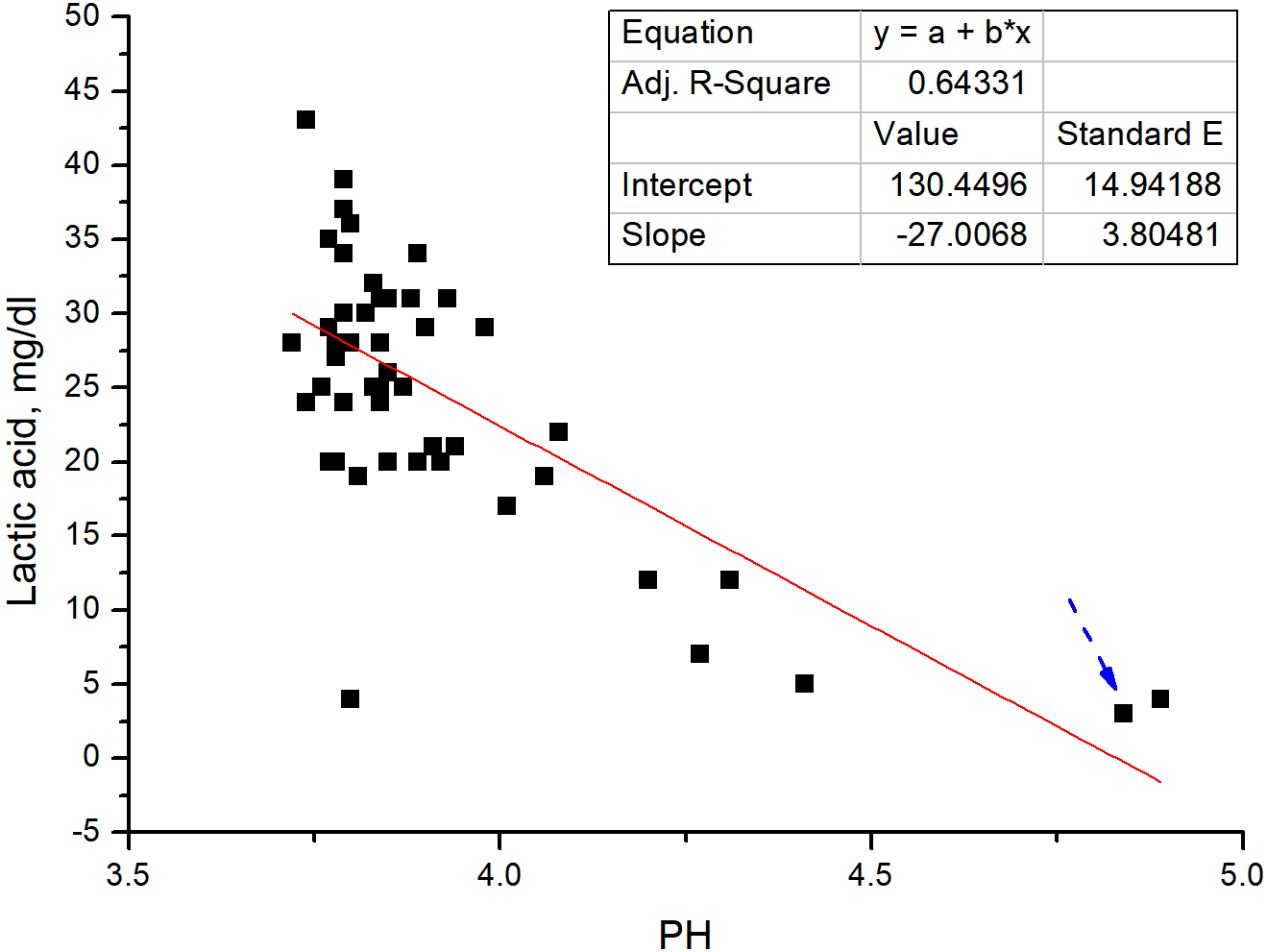
Ensiling technique is becoming a common way to preserve fresh corn and grass. This study aimed at evaluating the silages’ quality with a new type of inoculants designed based on the microflora information of the native silages. A total of 33 silage samples were analyzed for their microbial community structure by high throughput sequencing. A total of 73 dominant strains were isolated with a clear goal. The corn was ensiled by dominant strains (Lactobacillus buchneri II JC1-5: Acetobacter pasteurianus DHL-8 = 4:3; 109 CFU/g) as inoculants. In a 15-day trial period, the aerobic stability of this inoculated silage was significantly higher than the control group. There was a positive correlation between lactic acid content and pH/mold number (R2 values of 0.64331 and 0.48584, respectively). Compared to the control group, ammonia nitrogen (NH3-N), water soluble carbohydrates (WSC), neutral detergent fiber (NDF), and acid detergent fiber (ADF) decreased by 57.7%, 12.5%, 1.4%, and 7.5%, respectively; crude protein (CP) increased by 1.1%. Our results indicate that the new type of inoculants can improve the quality of the silage.
Keywords
silage
inoculants
microflora
high throughput sequencing
Funding
This work was supported without any funding.
Cite This Article
APA Style
Zhuang, X., Han, W., Xue, Z., & Miao, H. (2024). Evaluation of the Compound-microbes-preparation Based on Microflora Information as Inoculants in the Silage. Agricultural Science and Food Processing, 1(1), 29–37. https://doi.org/10.62762/ASFP.2024.516036
Publisher's Note
IECE stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Institute of Emerging and Computer Engineers (IECE) or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue