Abstract
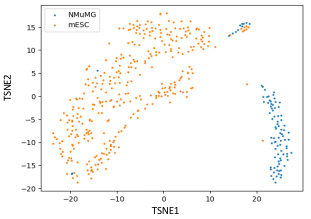
The volume and complexity of data in various fields, particularly in biology, are increasing exponentially, posing a challenge to existing analytical methods, which often struggle with high-dimensional data such as single-cell Hi-C data. To address this issue, we employ unsupervised methods, specifically Principal Component Analysis (PCA) and t-Distributed Stochastic Neighbor Embedding (t-SNE), to reduce data dimensions for visualization. Furthermore, we assess the information retention of the decomposed components using a Linear Discriminant Analysis (LDA) classifier model. Our findings indicate that these dimensionality reduction techniques effectively capture and present information not readily apparent in the original high-dimensional data, facilitating the visualization and interpretation of complex biological data. The LDA classifier's performance suggests that PCA and t-SNE maintain critical information necessary for accurate classification. In conclusion, our study demonstrates that PCA and t-SNE are powerful tools for visualizing and analyzing high-dimensional biological data, enabling researchers to gain new insights and understandings that are challenging to achieve with traditional approaches.
Funding
This work was supported by Name of funding agency under Grant 16JK1048.
Cite This Article
APA Style
Wang, Z., Zhang, P., Sun, W., & Li, D. (2024). Application of Dimension Reduction Methods to High-Dimensional Single-Cell 3D Genomic Contact Data. IECE Transactions on Pattern Recognition and Intelligent Systems, 2(1), 20–25 https://doi.org/10.62762/TIOT.2024.186430
Publisher's Note
IECE stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Institute of Emerging and Computer Engineers (IECE) or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue