IECE Journal of Image Analysis and Processing
ISSN: 3067-3682 (Online)
Email: [email protected]


Agriculture is a vital part of all economic systems and a basic source of food that ensures life will continue. To maximize output and raise quality, the agriculture industry must be improved. This entails creating the ideal environment for crops and plants to develop healthily. Plant degradation is typically caused by diseases. The Food and Agriculture Organization of the United Nations (FAO) estimates that the annual cost of these diseases to the world economy is around $220 billion. They cause harm to crops and, occasionally, their complete destruction. In fact, viruses, bacteria, fungi, and microscopic animals attack plants, altering their original form and affecting their important functions [1].
Plants can be saved when infections are detected early and neutralized. To protect crop health and ensure ongoing agricultural productivity, early diagnosis of plant diseases is essential. However, manual inspection is a major component of traditional systems, which makes them unreliable, time-consuming, and unsuitable for large-scale farming. Due to a lack of experience, farmers frequently find it difficult to detect infections early on, which causes delays in interventions and large crop losses. Given that agriculture is the foundation of many economies and food security systems, automated, precise, and effective disease detection methods are desperately needed. Deep learning models have emerged as a promising solution to these problems.
The crops are safeguarded, and losses are prevented to a greater extent the sooner they are identified. Due to a lack of expertise and experience, the conventional methods of disease detection—which are primarily reliant on human diagnosis—are insufficient and time-consuming [2]. The diagnosis must be based on a more trustworthy technique because the data collection method and verification frequency are also insufficient. Modern technologies have been provided as an automatic way to identify plant diseases for this purpose [3]. Among these technologies, advanced instruments like sensors, drones, and robots have transformed farmers’ field management [4], and machine learning has opened up new avenues for data analysis [5].
With encouraging outcomes, the use of deep learning and machine learning approaches in plant disease identification is a quickly developing subject [6]. However, because deep learning-based techniques rely on automatic feature extraction rather than human feature selection, they have demonstrated their efficacy above other machine learning techniques, particularly in relation to image identification [7]. DL-based methods for the Identification and detection of plant diseases have been proposed in a number of research studies. However, there are a lot of barriers that keep this technology from being used more effectively. The impossibility of gathering dataset photos for every illness across all leaf kinds is, in fact, one barrier. Furthermore, a number of illnesses are spreading quickly, making it difficult to catch them on leave on time.
Furthermore, the study [8] demonstrated that the system is impacted by both the crop’s and the disease’s properties during the learning process. This implies that features taken from one crop and illness cannot be transferred to other crops and diseases. Classification model training on a dataset comprising pairs of various crops and diseases is necessary to create a generalized model for classifying plant diseases. Unfortunately, there isn’t a dataset like that, and creating one is difficult, if not impossible. On the planet, there are millions of crops and plants, and millions of diseases could harm them.
Even though deep learning has demonstrated remarkable outcomes in the Identification of plant diseases, current models frequently lack generalization and resilience. The majority of methods are trained on certain datasets and have trouble detecting illnesses in new crops or in different environmental settings. This constraint occurs because small, disease-affected areas are not given as much attention to conventional models as the overall appearance of the leaf. Developing dependable, scalable solutions for actual agricultural settings is made more difficult by the lack of labeled datasets that reflect a variety of crop kinds and disease categories. To overcome these constraints, a model that can accurately identify diseases in a variety of crops and environments is needed.
This study presents a unique deep learning-based approach that aims to address this obstacle by generalizing the method for identifying plant diseases in various kinds of plants, hence overcoming the limits of specific crop training difficulties reported in the existing research. Our method is centered on the primary goal of diagnosing the disease, not just the sick leaf’s outward look. It places special emphasis on identifying the healthiness or illness in tiny leaf fragments. This study produced a method to achieve this, which involves dividing each image of leaves in the training dataset into smaller patches, so separating the disease from the healthy parts. By using this dividing approach, it is easier to extract disease-related information from the crops and enhance healthy data.
Afsharpour et al. [9] and Albahar [10] claim that DL models enhance fruit disease diagnosis and boost agricultural productivity. For the detection of fruit rot, the approach suggested in [10] was more reliable. By altering factors like lighting and picture obstructions, the suggested approach was able to overcome the consequences.
Huang et al. [11] proposed neural network enhanced multi-scale feature extraction and citrus fruit disease detection by combining the form of the inception module with the cutting-edge EfficientNetV2. The VGG was used instead of the U-Net backbone to enhance the network’s segmentation performance. The proposed neural network, which was developed by combining the Inception module with the cutting-edge EfficientNetV2, enhanced multi-scale feature extraction and citrus fruit disease detection. Lokesh et al. [12] proposed an artificial intelligence strategy for identifying leaf sickness that incorporates generative adversarial networks (GAN) and deep learning. The strategy surpassed current techniques, achieving higher classification accuracy for healthy and diseased leaf images. Wang et al. [13] improved the design of ResNet by suggesting a multi-scale ResNet. The system is used to identify vegetable leaf diseases on constrained hardware by lowering the model’s size. The multi-scale ResNet has outperformed other systems (VGG16, AlexNet, SqueezeNet, and ResNet50). Ren et al. [14] proposed a new system, the deconvolution-guided VGG network, for detecting tomato leaf illnesses and segmenting disease areas. The proposed model is capable of dealing with all image difficulties, including low light, shadows, and occlusion. Guo et al. [15] created a customized tomato leaf disease identification system for Android mobile devices. This method easily differentiates between different diseases at various stages by altering the steps of the AlexNet design, resulting in the Multi-Scale AlexNet.
Using an improved version of MobileNetV3, Qiaoli et al. [16] described a real-time, non-destructive method for detecting tomato illnesses. The method addresses the problem of model overfitting brought on by insufficient data by optimizing MobileNetV3. In order to differentiate between healthy and diseased leaves, Khalid et al. [17] concentrated on training a DL model, namely the YOLOv5 system. Both public and exclusive datasets were then used to test the trained system. The results show how quickly and precisely the model can detect even tiny disease areas on plant leaves. Panchal et al. [18] suggested a strategy for detecting crop diseases that includes segmenting images, labeling diseased leaves, pixel-based operations, feature extraction, and convolutional neural networks (CNN) for disease classification. To identify and segment tomato plant illnesses, Shoaib et al. [19] combined semantic segmentation methods (U-Net and Modified U-Net) with deep learning (Inception Net).
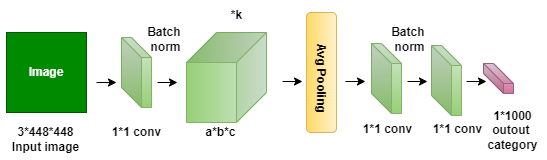
The suggested model, shown in Figure 1, creates four classes for assessing and estimating the suggested frame by utilizing four well-known transfer learning techniques: ResNet152, VGG19, DenseNet169, and MobileNetv3. After being subjected to four different transfer learning techniques, the data is analyzed and split into two sets: 80% for training and 20% for testing. This division is essential for generalizability assessment, model performance validation, and training. The suggested model shows reliability in a range of situations. In this work, we extend the dataset to train a deep learning model in plant disease diagnosis by the use of image augmentation, a crucial method utilizing Keras’ Image Data Generator. The model is exposed to a greater variety of variations by generating altered copies of images with rotations, zooming, and flipping, which enhances the model’s capacity to handle new data. This is essential to simulate the variability found in medical imaging and strengthen the model’s resistance to noise and fluctuations. The ultimate objective is to create a robust and dependable deep learning model, particularly in the medical field where data is scarce and case diversity and unforeseen conditions are critical. During model training, changes like zooms, flips, shifts, and rotations are added to help create balanced classes.
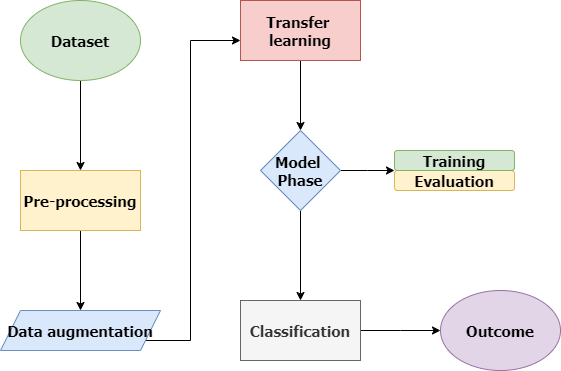
This augmentation technique helps create a training dataset that is more varied and extensive, which improves the model’s ability to generalize in a wide range of situations. There are two benefits to using ImageDataGenerator while training a model. In order to strengthen the deep learning model’s ability to recognize complex patterns and features, it first makes sure that the model is exposed to a wider range of training data. Furthermore, the automatic production of enhanced images improves the resilience of the model by reducing its susceptibility to overfitting and increasing its ability to adjust to a wide range of input variables. This augmentation-driven method has been shown to be effective in raising deep learning models’ overall performance, which improves accuracy and durability in practical applications [20].
A machine learning technique called transfer learning allows a model to be learned for one job and then applied to another that is related to it. This approach shortens the effort and time needed to create high-performing systems, particularly for challenging jobs like natural language processing and picture recognition. Through fine-tuning an established model’s weights, scientists can effectively address novel problems. By utilizing the knowledge gathered from a sizable dataset during the first training phase, transfer learning enables the system to efficiently handle novel difficulties.
This method differs with the usual way of training a system from the ground up, which can be resource intensive and time-consuming. In several fields, such as speech recognition, picture identification and natural language processing, transfer learning has shown promise, particularly in situations with little training.
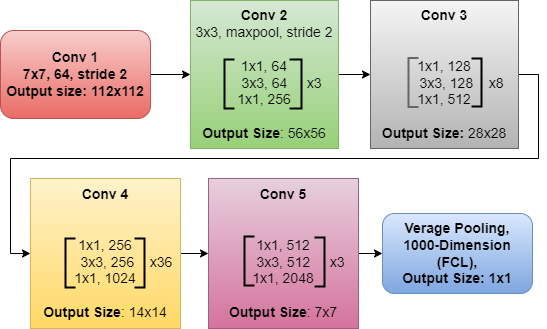
Microsoft Research created ResNet-152, DCNN architecture with 152 layers. The main advancement in it is the use of residual connections or skip connections, which allow the network to learn residual functions and ease the training of exceptionally deep networks [21]. ResNet-152 is effective at tasks like object recognition and image classification because of its depth, which enables it to excerpt complex patterns and features from data. When combined with skip connections, this architectural depth solves the vanishing gradient issue and makes it simple to train incredibly deep networks, as seen in Figure 2.
An advancement over the original VGG16 design is the DCNN (deep convolutional neural network) method called VGG19. There are sixteen convolutional layers and three completely linked layers, totaling 19 layers in this model. VGG19’s deep structure, which employs 3×3 convolutional filters for feature extraction, allows it to extract complex patterns and features from picture data [22]. By reducing the spatial dimensions of input, max-pooling layers minimize computing complexity. Due to the entire connectivity of the final layers, predictions derived from high-level characteristics taken out by the convolutional layers are possible. For nonlinearity, VGG19 employs the ReLU (Rectified Linear Unit) activation function. Often employed in image classification. In terms of efficiency and performance, it has been outperformed by more recent designs, such as ResNet and Inception, despite their simplicity and depth. Figure 3 shows the VGG19 architecture.
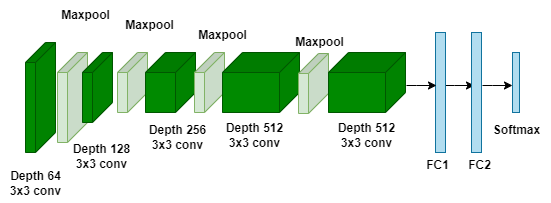
A CNN architecture called DenseNet169 was created to address issues with gradient flow and feature reuse in deep networks. In order to restrict the feature’s growth map and minimize its spatial dimensions, transition layers are used in between dense blocks. The Global average pooling layer, which lowers the number of parameters and improves expansions, is frequently used in DenseNet designs. DenseNet169 has proven to perform well in challenges involving image classification. The architecture of DenseNet169 is shown in Figure 4.
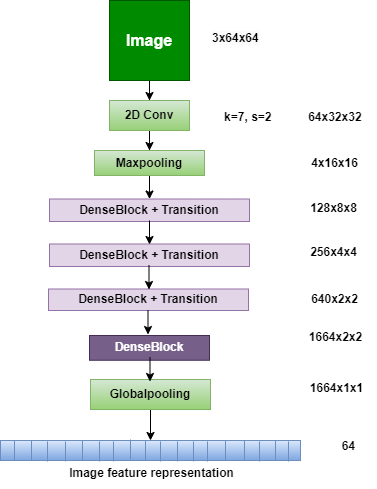
A neural network architecture called MobileNetV3 was created for edge and mobile devices with limited processing capacity. This third version of the MobileNet series prioritizes accuracy, efficiency, and speed. Two variations, MobileNetV3-Large and MobileNetV3-Small, lightweight inverted residuals and resource-efficient construction blocks are among the key features [23]. The architecture of MobileNetv3 is shown in Figure 5.
A particular dataset was used to train the Transfer Learning (TL) model, and a matching test dataset was used to assess it. Keras, Sklearn, and TensorFlow cooperate to ensure the model’s success. For high-end systems, 128 block size was ideal. Both the test and train sets were subjected to cross-entropy loss. The results are reported in Table 1. The DenseNet169 model behaved differently than the VGG19, MobileNetv3, and ResNet152 models, which all had close training and validation losses.
This study highlights how essential it is to use a Deep Learning model for the Identification of plant disease. 20% of each class was chosen at random from 14,059 photos in the PlantVillage dataset that were tested. The model classified leaves as either healthy or ill with an accuracy rate of above 95%. However, some photos were incorrectly categorized because of dirt or poor illumination.
The Adam optimizer was utilized in the study to train 50 models using cross-entropy loss. The results indicated that whereas DenseNet169 had a different pattern, with training loss reduced and validation loss increasing, VGG19, MobileNetv3, and ResNet152, models had near validation and training losses.
MobileNetv3 had the best testing accuracy of 99.75% among the four transfer learning models that were assessed. DenseNet169, on the other hand, displayed oscillations, suggesting sensitivity to changes in the dataset. The accuracy of ResNet152 was continuously high at 98.86%, but MobileNetv3 outperformed it due to its lightweight design and effective feature extraction. These findings demonstrate how crucial model architecture is for striking a balance between generalization, accuracy, and computational economy. VGG19 and ResNet152 models had training and validation losses, but MobileNetv3 demonstrated a steady training process with few oscillations. ResNet152 demonstrated the best performance among the four transfer learning models assessed in the study, with a validation loss of 0.0241 and 98.86%.
|
Testing | Training | |||
|---|---|---|---|---|---|
| Loss | Acc | Loss | Acc | ||
| VGG19 | 0.124 | 95.62 | 0.045 | 99.07 | |
| ResNet152 | 0.185 | 96.92 | 0.0603 | 98.86 | |
| MobileNetv3 | 0.127 | 98.52 | 0.0359 | 99.75 | |
| DenseNet169 | 0.958 | 97.53 | 0.0241 | 99.22 | |
DenseNet169’s training and validation accuracy fluctuated the most, despite having the lowest accuracy. Promising outcomes were also displayed by VGG19 and MobileNetv3, however there was some fluctuation in validation accuracy.
The suggested transfer learning systems use the confusion matrix to evaluate their results, which includes metrics such as recall, precision, accuracy, and F1 score. Using common criteria, such as accuracy, precision, recall, and F1 score, the model’s performance was assessed. Precision quantifies the proportion of identified diseased leaves that were correctly diagnosed, whereas accuracy indicates the overall correctness of forecasts. The F1 score offers a compromise between precision and recall, whereas recall shows how successfully the model detected every diseased leaf. The confusion matrix, which is commonly a square matrix, provides a thorough perspective of model outcomes. Table 2 shows the confusion matrix, with FP denoting false positives, FN denoting false negatives, and TP denoting true positives. The F1 score is calculated as the harmonic means of recall and precision.
This study evaluated the proposed model’s performance using multiple indicators and provided the findings. The model’s resilience is demonstrated by the low false positive and false negative rates, while the high true positive rates in every category show that it successfully detects a variety of disease types. The model’s capacity for generalization is further supported by its excellent performance across several classes. A useful tool for evaluating the model’s classification results is the confusion matrix. This matrix categorizes diseases from 1 to 3, with each integer identifier representing a specific disease type: 1 for "fungal diseases," 2 for "bacterial diseases," and 3 for "viral diseases." This systematic numbering clearly represents the model’s classification findings. The confusion matrix shows that the MobileNet model outperformed expectations.
These numerical values in the matrix offer important information about how well the model classifies photos of various conditions. The maximum accuracy was attained by ResNet152, which was followed by DenseNet169, MobileNetV3, and VGG19, which all showed respectable accuracy but fell short. These tests, which were carried out after each model was trained for 50 epochs, indicate that ResNet152 performed better than the other systems on a consistent basis. The four models’ different topologies highlight the complex trade-offs between system computational efficiency and accuracy. This study gives important insights into well-informed decision-making by helping to identify their strengths and limitations.
A thorough summary of the accuracy measurements for each model at both the highest and least epoch counts is given in Table 2. With the highest validation accuracy of 98.52% at epoch 45 and epoch 50, this study get the greatest training accuracy of 99.75%, and the MobileNet model performs better than any other model. The names Mi_Acc and Mx_Acc stand for Minimum Accuracy and Maximum Accuracy, respectively, and Mi_Ep and Mx_Ep for Minimum Epochs and Maximum Epochs, respectively, to aid in clear communication and notation.
|
Stage | M×-Ep | M×-Acc | Mi-Ep | Mi-Acc | ||
|---|---|---|---|---|---|---|---|
| VGG19 | Training | 50 | 98.78 | 1 | 70.22 | ||
| Testing | 50 | 96.91 | 1 | 81.45 | |||
| DenseNet169 | Training | 50 | 98.12 | 1 | 69.34 | ||
| Testing | 45 | 97.78 | 6 | 80.96 | |||
| ResNet152 | Training | 38 | 99.08 | 1 | 95.42 | ||
| Testing | 47 | 98.68 | 2 | 90.77 | |||
| MobileNetv3 | Training | 30 | 99.75 | 1 | 77.12 | ||
| Testing | 46 | 99.52 | 1 | 90.27 |
| Author | Year | Dataset | Method | Accuracy |
|---|---|---|---|---|
| Wang et al. [13] | 2020 | AI Challenge2018, Self, PlantVillage, | CNN (Multi-scale ResNet) | 95.95% |
| Ren et al. [14] | 2020 | PlantVillage | CNN (Deconvolution Guided VGGNet) | 93.25% |
| Guo et al. [15] | 2019 | PlantVillage, Self | CNN (Improved Multi-Scale AlexNet) | 92.7% |
| Qiaoli et al. [16] | 2022 | PlantVillage | CNN (MobileNetV3) | 98.25% |
| Khalid et al. [17] | 2023 | PlantVillage, self | CNN (YOLOv5) | 93% |
| Panchal et al. [18] | 2023 | PlantVillage | CNN (naïve network) | 90.40% |
| Shoaib et al. [19] | 2022 | PlantVillage | CNN (Inception Net) | 99.12% |
| Our proposed model | 2024 | Plant village | Transfer Learning Approach | 99.75% |
By juxtaposing the current state of the field with the anticipated outcomes of this research, Table 2 and Figure 6 comprehensively illustrate the evolutionary trajectory and projected advancements in plant disease detection and classification methodologies.
The potential for practical use in agricultural monitoring systems is indicated by the MobileNetv3 outstanding accuracy and stability. Farmers can use the model to detect diseases in real time scenarios using drones or mobile devices, allowing for prompt action, and minimizing crop loss. This useful tool can help farmers better control crop health, reducing yield loss and guaranteeing sustainable farming methods.
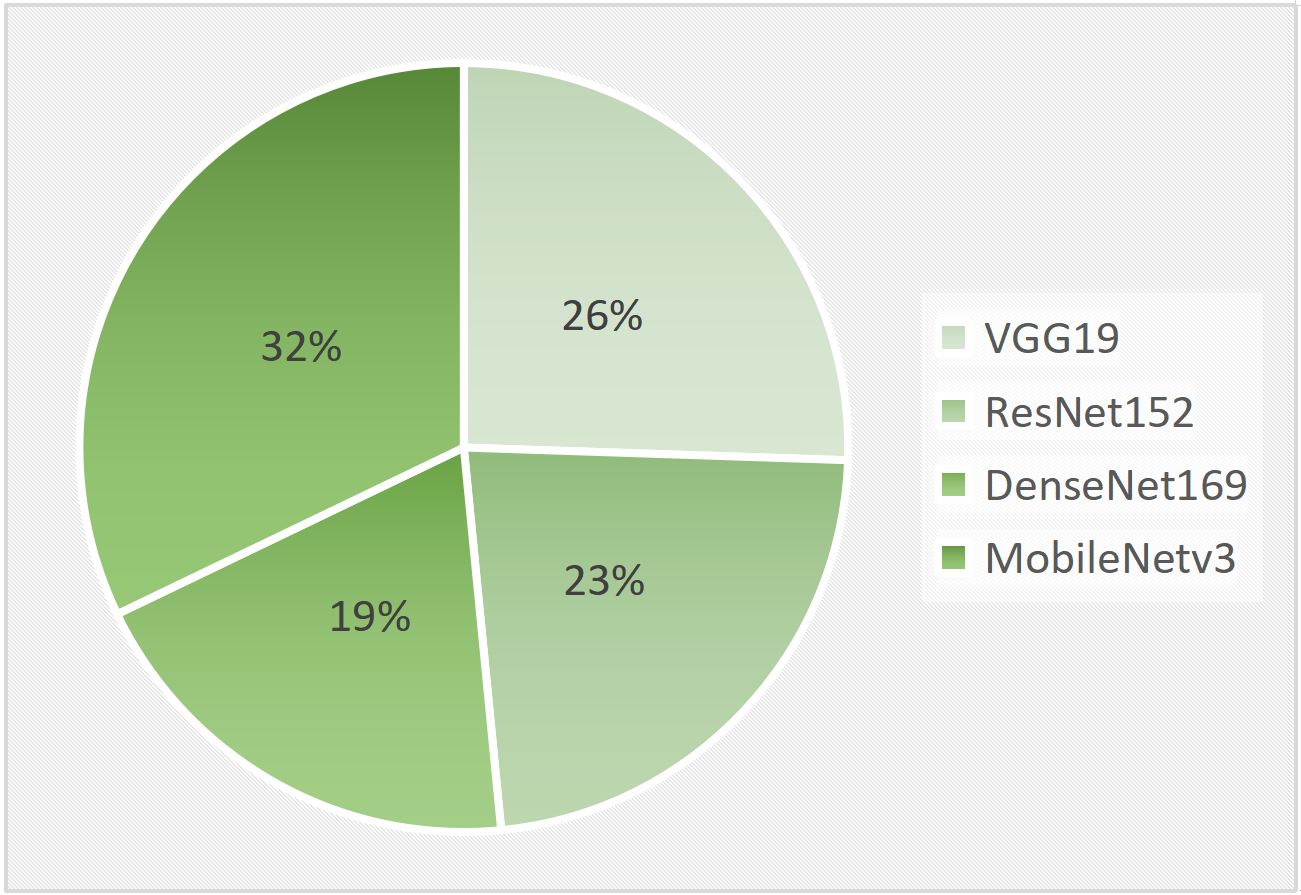
A crucial component of the proposed strategy is the MobileNetv3 model, which has an amazing 99.75% accuracy rate. This demonstrates that the model is a useful tool for disease diagnosis since it can effectively identify and predict the presence of any illness in plant pictures.
The comparison of classification accuracy between the suggested and other current models is shown in Table 3. The comparison of classification accuracy between the suggested and other cutting-edge techniques is shown in Figure 6. With an accuracy of 99.75%, Figure 7 makes it abundantly evident that the suggested system carried out better than other models.
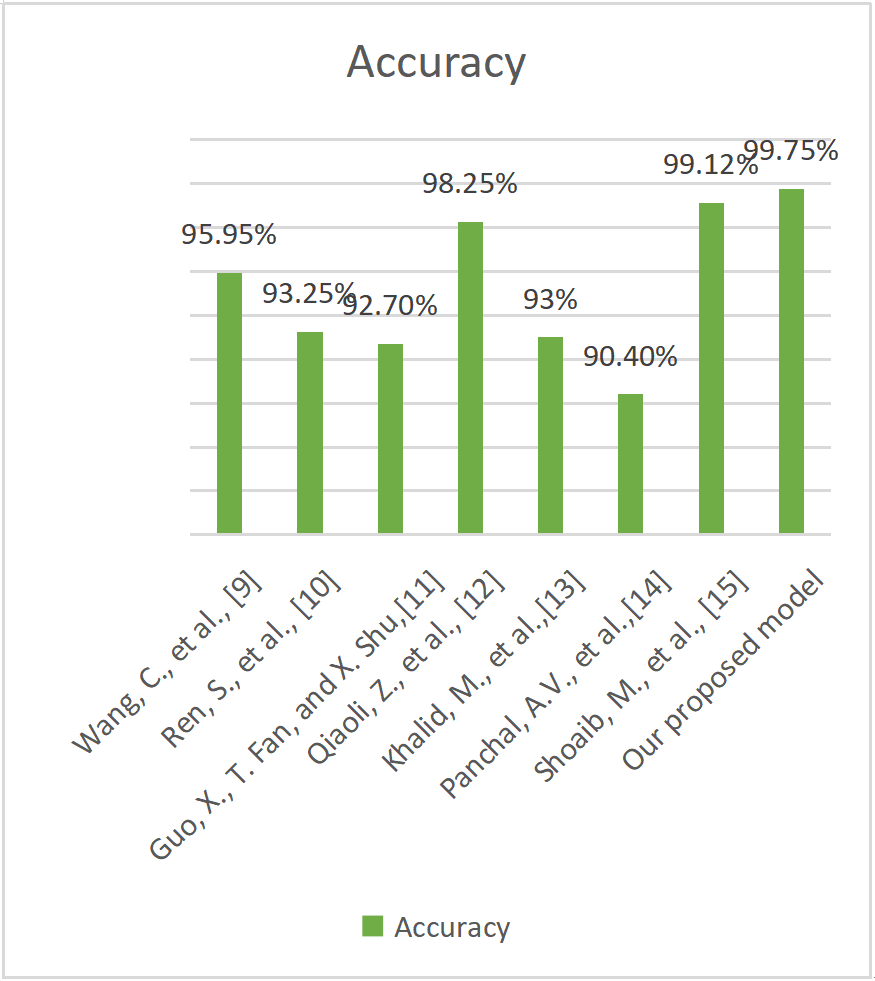
The comparison analysis shows that the suggested MobileNetv3-based model performs better than the current state-of-the-art methods for detecting plant diseases. Table 3 illustrates that the accuracy of the suggested method was higher than that of earlier models, including Multi-scale ResNet (95.95%), Deconvolution-Guided VGGNet (93.25%), and Improved Multi-scale AlexNet (92.7%). The MobileNetv3 model, on the other hand, outperformed all other techniques with an exceptional classification accuracy of 99.75%.
The model’s capacity to generalize across many kinds of crop and disease circumstances while preserving computational efficiency is what makes this work significant. The suggested approach concentrates on identifying discrete diseased patches rather than the overall appearance of the leaf, in contrast to standard models, which frequently struggle with changing environmental circumstances and emerging disease categories. This method improves the model’s resilience and qualifies it for use in practical agricultural applications, such as real-time field monitoring via drones and mobile devices.
Therefore, the suggested method is a useful tool for precision agriculture since it not only increases detection accuracy but also tackles critical issues with model generalization, computational effectiveness, and realistic deployment.
The work rigorously evaluates the effectiveness of four different transfer learning systems— VGG19, ResNet152, MobileNetv3, and DenseNet169, on the Plant Village Dataset. The evaluation includes critical performance indicators like precision, accuracy, recall, and f1-score. ResNet152 outperformed all other models in the study with an impressive accuracy of 98.5%. Furthermore, MobileNetv3 has outstanding effectiveness, with 99.75% accuracy, demonstrating its excellent results in plant disease detection. Integrating multiple imaging modalities enhances the proposed model’s performance, durability, and real-world application. Even though the suggested model performed remarkably well on the Plant Village Dataset, still there exist a number of validity risks. The dataset’s controlled conditions may have contributed to the high accuracy attained, and the model’s performance may differ in real-world settings with varying lighting, background noise, and leaf occlusions. Furthermore, the research’s ability to be applied to different agricultural contexts was limited due to its reliance on a single dataset. Early signs that manifest on other plant components, like stems and fruits, may also be missed due to the emphasis on leaf-based disease diagnosis.
Future research will concentrate on adding real-world photos from various agricultural settings to the dataset, combining RGB images with multimodal imaging (thermal, hyperspectral, and infrared), and refining the model for deployment on edge devices like smartphones and drones for real-time monitoring in order to overcome these limitations. Future study could look into expanding the suggested model’s usage improving its flexibility. This extension has the ability to expand the influence of models. To summarize, the proposed approach, notably MobileNetv3 and ResNet152, demonstrate significant potential for improving image classification. Further research into various imaging modalities can provide vital insights and improve illness detection applications significantly. Future study will evaluate the efficacy of these models in actual situations using various datasets. The primary objective of this study is to refine image enhancement techniques specifically designed for particular disease groups, thereby facilitating the development of an optimized computational model and a comprehensive, balanced dataset for enhanced diagnostic applications.
 Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 
Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/iece/