Agricultural Science and Food Processing
ISSN: 3066-1579 (Online) | ISSN: 3066-1560 (Print)
Email: xucm@btbu.edu.cn


Genomics is an interdisciplinary biological discipline that involves the collective characterization and quantitative study of all genes in an organism and the comparative study of different genomes, rather than the structure and function of a single gene [2]. As a data-driven science, it mainly uses machine learning to capture the dependencies between data and deduce new biological hypotheses [1].
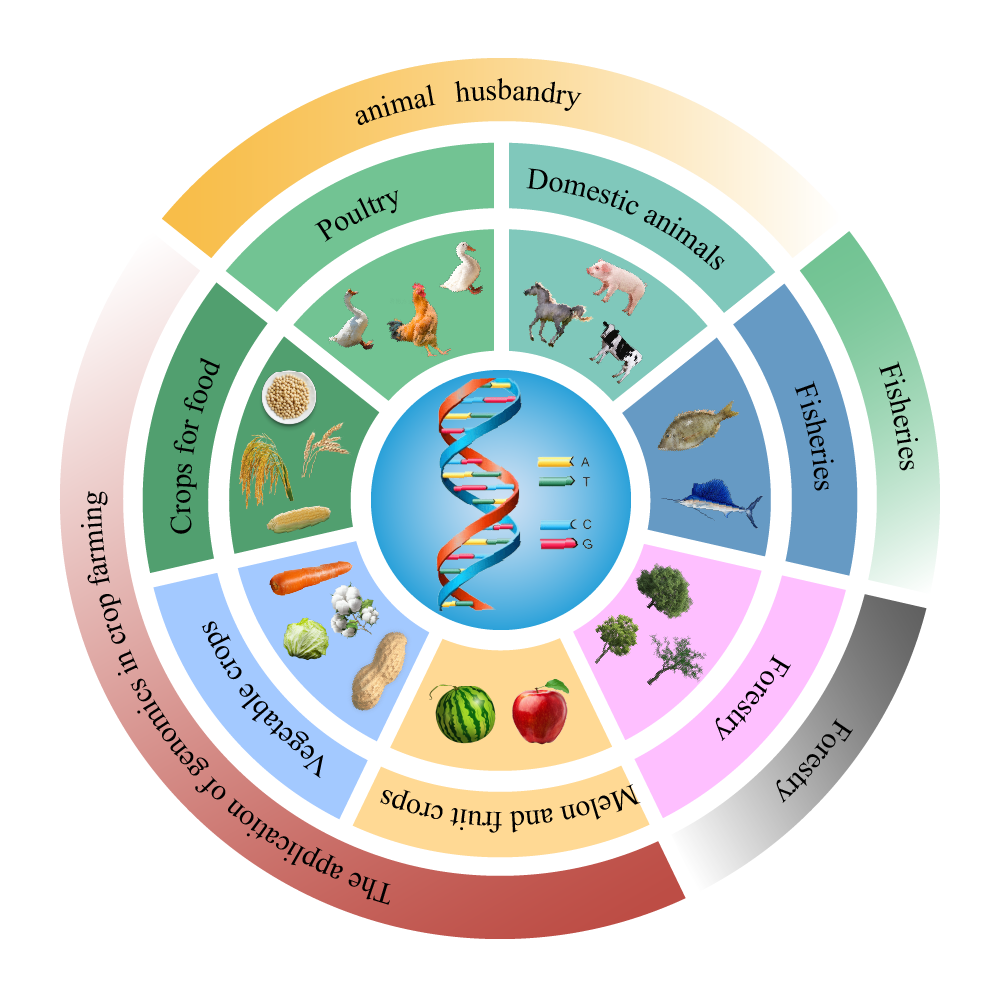
In the context of a growing global population, the urgent problem for agricultural development is: how to produce more food with limited resources, while having less impact on the environment [3]. The human population is expected to reach 10 billion by 2050, but crop yields appear to be leveling off or even declining due to climate change and limited supply of arable land, with a 60 percent increase needed to feed the world's 10 billion people. Therefore, improving agricultural productivity and sustainability is critical for the world. There is an urgent need for continuous scientific breakthroughs and technological innovations to ensure global food security in the future [4]. Genomics is of great significance in breeding, animal husbandry and sustainable agriculture. Genomics can make great contributions to plant crop improvement and animal breeding improvement by intercrossing with computer science and other disciplines. At the same time, genomics can also improve the adaptability of crops to the environment, respond to climate change, and ensure global food security by developing sustainable crops that can adapt to environmental changes [5]. As shown in Figure 1, genomics technologies such as whole-genome sequencing and genome-wide association studies (GWAS) have enabled the identification of key genes and quantitative trait loci (QTLs) associated with agriculturally important traits, leading to the development of crops with enhanced nutritional value, yield, and stress resistance. These advances demonstrate the power of genomics in facilitating precision breeding and sustainable agriculture to address global food security challenges.
Rice is an important cereal and model plant.Rice genomics research can be divided into three approaches: structural genomics, functional genomics and quantitative genomics [2]. Due to the complexity of genomics research, two approaches are commonly used in combination. The combination of structural genomics and functional genomics can comprehensively analyze the composition, structure and function of genes. Yang et al. [11] used the rice seedling HuizhanNano as the experimental material and found that Pi2, Pi2, Pi2, and Pi2 genes play a key role in the strong disease resistance of huizhan using nanopore long-read technology. Using functional molecular markers, Xuan et al. [8] found a significant correlation between the blast resistance gene Pita and blast resistance, and proposed to extend the lifespan of resistant varieties by designing reasonable distribution and rotation of blast resistance genes.
The key advantage of integrating structural and quantitative genomics lies in precisely analyzing complex trait-genome associations through genomic physical structure and quantitative variation. Das et al. [9] has enriched the genomics of flower organs in rice crops by using high-throughput whole genome sequencing technology, pre-processing and mapping of reads, and variation analysis. Liu et al. [10] used 180 rice germplasms with large yield variations as experimental materials to perform association analysis of genes related to grain weight by target sequence capture sequencing, and found that TGW-HC4 was the best haplotype combination. By using genome resequencing technology, Yu et al. [3] identified a large number of genes and genomic regions or QTL controlling green traits, which enabled green super rice to be breeding and promoted.
The integration of functional and quantitative genomics enables systematic gene function analysis and precise quantification of genetic effects. Lee et al. [6] combined QTL mapping and GWAS analysis techniques to analyze heading date genes and rice nutrition-related genes, respectively, and showed that heading date genes had pleiotropic effects on grain, and accelerated heading and nutrient-rich rice breeding could be achieved by locating mutations and mutagenesis. The application of genomics has improved the nutritional content of rice[6, 7].
Beyond integrating these approaches, alternative avenues for investigation exist.By reviewing the origin and domestication history of African rice, Wambugu et al. [12] clarified that the progress of genomics could continuously change the previous understanding of rice domestication, so as to had a more accurate understanding of rice domestication process from the genomic level, which can eventually guide crop improvement. Ahmad [13] reviewed the use of several genomic and transcriptomic approaches to understand the ability of rice to withstand abiotic stress. Zhang et al. [14] provided the eRice database as an online research platform for the rice research community. To provide its epigenomics and genomics resources and will continue to enrich epigenetics data. In summary, genomics advances rice breeding through enhanced nutrition, accelerated heading, and improved blast resistance.
Wheat is the world's largest cultivated and most widely distributed food crop [15]. Modern biological techniques have been widely used in wheat breeding [16]. Firstly, molecular marker technology has been widely used in wheat breeding. Kumar et al. [17] and Shewry et al. [18] used molecular marker technology to predict genotype and improve nutrient content of Indian wheat. They introduced the trait by identifying QTLS for dietary fiber content and created wild and weed relatives of wheat. Babu et al. [19] performed a cis-genic approach by molecular marker-assisted selection (MAS) to identify multiple anti-rust genes simultaneously.
Secondly, the technology related to sequencing is more and more emerging. Kayess et al. [20] used next-generation genome sequencing to conduct extensive gene screening for heat-tolerant wheat varieties, and combined phenomics and genomics to develop new varieties with high temperature tolerance. Li et al. [16] proposed that the IWGSC RefSeq v2.0 (the second version of the wheat reference sequence) had a nearly complete wheat genome sequence, which could isolate and characterized the genes for agronomic traits important for wheat molecular breeding in the future. Following this, Wang et al. [21] focused on understanding the origin and evolutionary history of wheat, applying resistance gene enrichment sequencing and k-mer-based association mapping markers to clone disease-causing genes and accumulate genomic data. The evolutionary history of wheat was revealed by genome analysis of wild and domesticated wheat [22]. In 2023, Sun et al. [23] reviewed the extensive application of MAS, QTL mapping, and GWAS in genomics-assisted breeding(GAB) in breeding programs. Thereafter, Roychowdhury et al. [24] reviewed methods to improve key agronomics traits such as yield, disease resistance, drought tolerance, and nutritional quality of wheat through precise genetic modification. Subedi et al. [25] performed a translational genomics study of grain size regulatory genes in wheat using the rice candidate gene OsGS3 through the candidate gene approach. Boehm et al. [26] focused on foreign introgression during wheat improvement, developing a genomics-based chromosome engineering pipeline to improve the efficacy and throughput of foreign introgression. Rhodes et al. [27] put forward the urgent need to use the global genomic surveillance system to continuously track and monitor wheat blast and prevent the global spread of rice blast. Finally, several important wheat genomics-related platforms are proposed,including WheatOmics[28], Online database of single grain wheat [29]. Sehgal et al. [30] developed WheatOmics, a multi-omics platform enabling wheat functional genomics research through gene identification, transcript analysis, regulatory mapping, gene networks, mutant/variant analysis, optimized for non-bioinformaticians.
To overcome the challenges and ensure the sustainable increase of maize yield, genomics-based breeding is required [31]. First, gene sequencing has made a major contribution to the elucidation of various mechanisms. By combining PacBio Sequel and Illumina HiSeq sequencing, Wang et al. [32] assembled a more continuous genome of Bipolaris maydis, providing a valuable resource for studying leaf dieback disease in southern maize and helping to elucidate the molecular mechanisms of its pathogenicity. After further high-throughput sequencing(NGS), Farooqi et al. [33] aimed at maize abiotic stress tolerance traits, combined with phenotypic analysis, and compared gene expression and function under extreme conditions to understand the molecular mechanism of abiotic stress in maize. QTLS for cold tolerance, heat stress, salt stress, and drought were also mapped. Subsequently, based on long-read sequencing technology, Liu et al. [34] used maize as a model to achieve gap-free genome assembly. This can eliminate uncertainty between genes and their regulators and understand the internal structure of repetitive domains, thus laying the foundation for epigenome analysis and ultimately creating seamless telomere assemblies of complex genomes in the future. Yang et al. [35] used maize HapMap and genomewide CRISPR mutation libraries to provide insight into population structural variation and the fact that a subset of transposable elements are transcriptionally active and can be induced under stress conditions, and developed an editing platform for high-throughput gene targeting in maize. This will revolutionize maize functional genomics research. Hu et al. [36] used genome assembly and population genome analysis to identify candidate genes associated with sweet maize traits. Dang et al. [37] has conducted GWAS and genomic prediction analyses for plant structural traits. It is concluded that the common origin of modern sweet corn in the United States was northern Mexico, and the plant configuration characters of sweet corn and waxy corn were understood.
For the GWAS technique, Waqas et al. [38] combined it with QTL mapping to explore important chromosomal regions and genes in maize under drought and heat stress. Farooqi et al. [33] reviewed the progress of maize multi-omics in abiotic stress resistance and breeding, highlighting the application and potential of genomics to metabolomics in stress resistance research. Using GWAS and co-expression analyses of root traits, Wu et al. [39] revealed genetic differences between aboveground agrtural traits and RSA traits and provided an important candidate gene resource for future maize root genetic improvement. Talukder et al. [40] targeted breeding of waxy maize by recessive wx1 introspection using three superior self-bred maize cultivars HKI323, HKI1105, and HKI1128. This maize has a shorter reproductive cycle, is rich in amylopectin, and can be used as a wx1 gene donor in breeding programs. Muntean et al. [31] outlined the application of approaches such as cis-genetics and epigenetics to enable maize breeding to move from domestication to precision breeding through assisted selection processes or precise breeding programs, and enable the development of new maize crops with high yield and quality performance as quickly as possible. McMillen et al. [41] targeted maize in the African region and used an ecophysiological model to disentangle the relationship between genes and the environment by integrating genomic information with physiology.
Finally, Woodhouse et al. [42] proposed MaizeGDB, a genomic database that can manage pan-genomic data, so that researchers can easily understand the structural and functional differences between maize and its direct relatives. Recently, Zhou et al. [43] proposed a method to dwarf maize. Based on the geneticrelationship between d024, indel-4 and ZmCYP90D1, they proposed to breed a new type of maize dwarf germplasm by crossing the corn-teosinte - Gramineum allopolyploid.
In recent years, significant progress has been made in soybean genomics, revealing the role of epigenetics in soybean regulation, polyploidy and diploidization, and the 3D structure of the soybean genome [44]. Sun et al. [45] reviewed various aspects of soybean genomics, including the application of 3D genomics, which was very extensive and comprehensive. More importantly, the important breeding trait of soybean is the relationship between oil yield and protein content, and many scientists have made great efforts in this regard. Liu et al. [46] used resequencing and GWAS to reveal key loci for soybean protein content and oil production. Kumar et al. [47] applied low-coverage resequencing, genome selection (GS), QTL mapping, and GWAS to seek ways to improve soybean seed oil and protein simultaneously. Kim et al. [48] identified candidate genes associated with protein, oil, and amino acid content in 203 wild soybean germplasm by GWAS and detected Single Nucleotide Polymorphism(SNP) markers for protein and oil content at loci on new chromosomes 15 and 20. Jia et al. [49] used SLAF-seq data for genome-wide association analysis, combined with transcriptome expression data, to identify three potential candidate genes with excellent haplotypes, These included Glyma.03G186200, Glyma.09G099500 and Glyma.18G248900, which facilitated potential MAS and improve oil production. In addition, for other important agrologic traits, Liu et al. [50] conducted individual de novo genome assembly of 26 representative soybean samples, constructed genome maps, and conducted pan-genome analysis, which revealed many new genetic variations and helped to find the correlation between genetic variations and candidate genes. Du et al. [51] proposed that genomics has played an important role in soybean resistance to biological stresses of Staygreen syndrome, soybean Mosaic virus, soybean cystinea, soybean common cutworm, Phytophthora soei root rot and stem rot using GWAS, linkage analysis and QTL analysis. Bhat et al. [52] introduced the application of GAB in soybean improvement. Through QTL analysis, high-throughput genotyping and high-throughput digital phenotyping (HTP) platforms, the utility of GAB in soybean has been greatly increased. By combining CRISPR-Cas9 and cutting-edge sequencing technologies such as RNA-seq and GWAS, Razzaq et al. [53] have made precise alterations of genetic information to greatly improve important traits such as yield, nutritional value, and stress resistance. Johnson et al. [54] reviewed the advances in genomics before and after soybean improvement in the last decade.GWAS and GS were proposed to improve the utilization efficiency of micronutrients in soybean and solve the problem of micronutrient deficiency in global soil.
Finally, the application of database is also indispensable. Razzaq et al. [53] also proposed the Soybean Genetics and Genomics Database (SoyBase) with extensive soybean genomics data. The database updates and new tools and features of SoyBase were detailed later by Brown et al. [55].
Peanut is an important food, oilseed and feed crop [56]. The development of sequencing technology has brought great progress to peanut genomics. Li et al. [57] used Chinese peanut as experimental material and obtained the first draft of genome sequence of Bacteroides peanutiae strain YY187 isolated from leaves by PacBio Sequel sequencing technology. Contribute to understanding the genetic diversity of fungal pathogens and further comparative genomics. Bhat et al. [56] performed whole-genome resequencing on 179 peanut samples to reveal the genome-wide structural and functional characteristics of SNP. In 2024, Raza et al. [58] proposed that GAB combined with NGS methods, high-throughput trait related markers, HTP, MAB, and GS can be used to design new varieties faster and help de novo domestication. In 2024, using long read sequencing and RNA sequencing, Xue et al. [59] introduced Amon2.0, a higher quality, more continuous, complete and accurate monticola genome assembly method, which was helpful for future molecular-assisted breeding. Ma et al. [60] used tetraploid peanut (Arachis Tifrunner) as experimental material to construct genomic Simple Sequence Repeats(SSRs) and combined with computer e-PCR analysis to enrich the genomic data of peanut SSRs . Tang et al. [61] used qRT-PCR analysis and CRISPR/Cas9 technology to identify a total of 19 AhFatB genes for improving peanut oil quality.
The impact of external stress on peanut development cannot be ignored. Huang et al. [62] conducted genomic research on peanut biological stress, and summarized the response of peanut to a variety of pests and microbial pathogens and the progress of omics. For example, Marker-Assisted Selection (MAS) and markassisted backcross have been successfully applied in peanut. O-methyltransferases (OMTs) play important roles in biological metabolism and stress tolerance. Cai et al. [63] conducted a genome-wide study of OMTs to provide a theoretical basis for understanding peanut development and stress response. For the Virginia peanut population, Newman et al. [64] used PCR allele competition expansion detection method, GS of high-quality markers and molecular marker assisted selection (MAS) methods to conduct a specific study on its genomic data. Wang et al. [65] reviewed that omics, especially genomic data, plays an important role in understanding the molecular basis of peanut seed development and also contributes to understanding the genetic and epigenetic mechanisms of key agronomic traits.
Cotton is an important fiber crop and model system in the world [66]. Firstly, Manivannan et al. [67] comprehensively reviewed the progress of cotton genomics in recent years, including the introduction of functional genes in cotton organelle genome and methods to improve cotton fiber quality, etc. In addition, the application of NGS, GS, 3D genomics and cotton pan-genome in cotton was introduced. However, Nadeem et al. [68] introduced the application of sequencing technology to cotton leaf rolling virus resistance and construction of mutation libraries induced by EMS (a chemical mutagen in breeding) in cotton, respectively [69]. Comparative genomics, developed by sequencing technology, has also been widely used. Using four Bacillus strains as experimental materials, Liu et al. [70] performed comparative genomic analysis and found that Bacillus velezensis was more abundant in secondary metabolites, which could effectively control verticillium wilt in cotton. Cohen et al. [71] used genome-wide comparative techniques to identify several nematode resistance genes, which in turn could be introduced and stacked into several upland cotton lineages. A more comprehensive understanding can be obtained by combining various genomic technologies, such as QTL, GWAS, RNA interference (RNAi) and virus-induced gene silencing, and 3D genomics. Naoumkina et al. [72] review the application of these techniques to improve both cotton fiber yield and quality. Iram described the application in drought tolerance of cotton [73]. Wen et al. [74] revealed the origin of cotton species and applied to the study of candidate genes related to fiber development, Yang et al. [75] reviewed future plans for breeding "super cotton". Wang et al. [76] used Shixiya No.1 (SXY1), an Asian cotton plant, as experimental material to identify and regulate candidate genes, which promoted the progress of cotton functional genomics. Shen et al. [77] obtained genomic data of 890 upland cotton varieties through genome-wide selective scanning detection, and analyzed that Introgression is affected by both artificial selection and environmental factors, which contributed to the understanding of the mechanism of intraspinal penetration.
Finally, the establishment of a database is very important for the study of cotton's association with genomics technology. In 2020, Yang et al. [78] proposed that a global data exchange platform should be established. After that, Zhang et al. [79] introduced a Gossypean genome database and detailed its functions and usage, such as the ability to search for orthologous gene ids. Liu et al. [80] constructed Fingerprint Finder by short-read sequencing, indicating that GWAS and fingerprint sites complement each other, which can more comprehensively understand the population genomics of cotton.
Chinese cabbage is a nutritious and economically significant vegetable crop [81]. Chinese cabbage can also be used as a model crop [82]. Ma et al. [82] proposed that high-density Chinese cabbage genetically mutated populations (CCGMPs) could promote the progress of functional genomics and use Chinese cabbage as a model to explore the function of leafy vegetable crops. Recent advances in the search for the cabbage genome, Lv et al. [83], based on High-throughput/resolution chromosome conformation capture (Hi-C) 's technology, assembled a high-quality draft cabbage genome. Xu et al. [84] discovered the importance of genomic SSRs markers for genetic linkage analysis, MAS, and genetic mapping by mining a large number of potentially variable SSNS through genome-wide bioinformatics surveys. As a kind of natural functional pigment, carotenoids are very nutritious. Cao et al. [81] used comparative genomics to study carotenoid biosynthesis genes, which laid a foundation for understanding the pathway of carotenoid biosynthesis and breeding cabbage with high carotenoid content. Yin et al. [85] used genome-wide data and bioinformatics methods to perform gene structure analysis, cis-acting element analysis, and collinearity analysis of the HD-Zip gene family, and found that the HD-Zip gene family has a regulatory role in carotenoid content and heat stress tolerance. In addition, for other agronomic shapes of cabbage, Ji et al. [86] used population comparative genomics, RNA-seq analysis, and qRT-PCR experiments to identify six genes that can be used for MAS and also understand the mechanism of heading traits. Wang et al. [87] combined multiple genomic data to analyze the glucosinolates (GSLs) biosynthetic genes, which helps to understand the synthesis mechanism of GSLs and prepare for the breeding of cabbage with high GSLs. Xing et al. [88] comprehensively analyzed the pseudo-response regulators(PRR) genes of cabbage cultivar 'BoZG17-1' through multiple sequence alignment PRR gene, gene structure analysis and cis-regulatory element analysis. It is beneficial to the cultivation of resistant varieties. Xu et al. [89] used genome sequencing, complete sequence alignment, genome structure comparison and phylogenetic analysis to study the cabbage chloroplast genome, which provided high-quality genetic breeding information. Chen et al. [90] studied the genome of Ogura CMS cabbage by gene sequencing, collinearity analysis and PCR amplification, and based on this, a new non-breeding cabbage, Bel CMS cabbage, was constructed. It has the advantages of less exogenous cytoplasm and no dead buds at the beginning of flowering. Finally, databases can provide rich genomic data, and Li et al. [91] introduced the content and functions of a newly developed database, the non-heading Chinese cabbage database, which will be combined with bioinformatics to facilitate comparative genomic analysis in the future.
Carrot is a nutrient-rich vegetable [92]. First, in order to improve the breeding of carrots, Macko-Podgorni et al. [93] used open-pollinated carrots as experimental materials and found that the expression of DcDCAF1 could regulate storage root development through GWAS. Liu et al. [95] improved hybridization of carrots using MAS, introgression, and third generation sequencing. Duran et al. [94] used sequencing genotyping and probe analysis to construct a genetic map of fertility restoration genes . Second, comparative genomics was also important. Zhao et al. [96] performed NGS and analysis of Aspergillus rhizobium isolate CBR2 (the causative agent of black rot of carrot) and Alternaria dauci (the causative agent of Alternaria leaf blip), respectively. Advances in comparative genomic analysis of carrots have been promoted. Zhe et al. [97] identified candidate genes for the formation of trichomes by resequencing and qRT-PCR analysis, which identified 12 genes associated with trichomes. Hao et al. [92] elucidated that DcNAC genes in carrot respond to abiotic stress by multiple sequence alignment and phylogenetic tree analysis. Finally, Rolling et al. [98] presented the content and functionality of the comprehensive database CarrotOmics, such as its ability to provide a visual carrot reference genome and drive genome-assisted selection.
Apple is a fruit of economic importance [99]. Many attempts had been made to address abiotic stresses. Liu et al. [100] used the 'Baleng Crab' cultivar as the experimental material, and developed two markers, SNP182G and SNP11G/SNP761A, to increase the salt-alkali tolerance of apple rootstock through SNP marker analysis and accurate prediction of salt-alkali stress damage index. Yao et al. [101] used "Golden Delicious" as experimental material to perform genome-wide identification and construct phylogenetic relationships for the HSP20 gene family, and finally identified several HSP20 genes as candidates for heat stress, which could contribute to the development of heat-tolerant apple varieties. Verma et al. [102] reviewed the role of multiple gene families in drought and salt tolerance in apple and introduced some miRNA candidates under investigation. In addition to abiotic stresses, pests and diseases are also a serious threat to apples. Anthrax is the causative agent of apple bitter rot. Khodadadi et al. [103] performed phylogenetic analysis and secondary metabolic gene cluster analysis by whole genome sequencing combined with bioinformatics software such as fastANI, which provided valuable genomic resources of anthrax and was beneficial to the disease management of apple stash. Carneiro et al. [104] completed the draft genome sequence of the pathogen Colletotrichum chrysophilum by sequencing the DNA of the chrysophilum strain M932 using a two-end sequencing system. It helps to develop varieties that can resist renal leaf spot disease and bitter rot disease. In order to select high-quality varieties, Shen et al. [105] proposed a new bioinformatics tool, bulked segregant analysis tool for out-of-crossing species and a genomics aided prediction model, they had used it to predict several QTLS and candidate genes for key agrologic traits in apple. Kumar et al. [106] used Malus domestica and wild Malus germplasm as experimental materials to study the genetic relationship of various nutrients in apple, including ascorbic acid, through apple 20 K SNP array genotyping, combined with GWAS and metabolite epitope analysis. This facilitates targeted breeding to improve different nutrients. Keller et al. [107] used apple REFPOP as experimental material and used FruitPhenoBox for gene phenotype analysis and SNP genotyping to study the genetic basis of apple shape and color, which helps to select the appearance of apple when breeding. Chen et al. [99] reviewed the previous research on red meat apples and proposed that MdMYB10 was an important research direction for the selection of high-quality red meat apples.
Finally, apple genomics data and research methods are also constantly enriching. For The 'Red Fuji' apple, Peng et al. [108] used HiFi long reads and Hi-C reads to analyze the high-quality haplotype 'Red Fuji' apple genome, which was a valuable resource for comparative genomics. In 2021, Minamikawa et al. [109] used 184 apple apple breeding lines as experimental materials to show that combining the data of infinium and the GRAS-Di genotype system, GWAS and genomic prediction(GP) can produce more accurate results. Then in 2024, Minamikawa et al. [110], using seven founders as experimental materials, through SNP genotyping chip system, GP and GWAS methods, the breeding value of the founder haplotype was revealed and the accuracy of these methods was evaluated.
Watermelon is the third largest fruit crop in the world [113]. Understanding the genetic changes in watermelon domestication history helped modern watermelon breeding scientists to generate their seamless genomes. Using 27 watermelon and its relatives as experimental materials, and found that watermelon was derived from Citrullus lanatus subsp. Cordophanus , which was helpful for analyzing good shape and introducing it into breeding. Renner et al. [112] used Illumina sequencing and other technologies to perform chromosome level genome assembly of Kordofan melon and found a large number of genome structural variations(SV), which can provide insights into genetic changes in watermelon domestication history and contribute to modern watermelon breeding. Yang et al. [113] used Citrullus lanatus cultivars to study the genetic map of watermelon by Perfect SNP, Target SNP-seq technology and construction of phylogenetic trees, which helps to promote the diversity of watermelon varieties. It helps to promote the diversity of watermelon varieties. In order to breed superior varieties, Yang et al. [114] used two watermelon mother species "1061" and "812" as experimental materials, and found the structural and functional properties of ClPRX gene family, such as its involvement in watermelon skin lignin synthesis, through qRT-PCR analysis and combined with multiple bioinformatics analyses. Yang et al. [113] used bulk segregant analysis Pipelines and RNA-Seq methods to identify important QTLS that could contribute to the development of drought-tolerant watermelon cultivars. Maraga et al. [115] used the hybrid progeny of BIL-53 and IIHR-140-152 as experimental materials, and identified two potential candidate genes for watermelon epidermal traits through QTL mapping and mapping, candidate gene analysis and sequencing, and SNP identification, Named Cla97C09G175150 and Cla97C09G175170. Mashilo et al. [116], for C. lanatus. The comparative analysis of traits, genetic characteristics and genomes was helpful for gene integration breeding.
Finally, some watermelon-related databases have gradually emerged. Deng et al. [111] used four watermelon germplasm as experimental materials to perform Genome de novo assembly and Oxford Nanopore Technology long-read sequencing. High-quality watermelon reference genomes and mutation libraries were obtained. Patel et al. [117] summarized the history and mechanism of watermelon anthracnose and proposed that for sustainable cultivation of watermelon anthracnose resistance, it is necessary to establish resistance gene libraries to continuously improve resistance. The application of genomics in other crops is also summarized in Table 1.
| Subjects | Technology | Conclusion | References |
|---|---|---|---|
| Lentils | NGS, MAS, SNP | Genomics for the promotion of lentil breeding, yield improvement, and adaptation to adverse environments. | Kumar et al. [118] |
| miRNAomics, epigenomics, phenomics | Developing Saline-Smart Lentils. | Ali et al. [119] | |
| CRISPR/Cas9, QTL | Review of drought tolerance mechanisms in lentils. | Saini et al. [120] | |
| genotyping,GWAS, SNP | The source of key genes responsible for salt tolerance in lentils was identified. | Dissanayake et al. [121] | |
| disease resistance breeding, hybridization, mutagenesis, molecular markers | This review summarized recent advances in improving disease resistance and climate adaptation of lentils. | Roy et al. [122] | |
| weeds | Genome Sequencing | Integrating multi-omics data can provide a comprehensive understanding of weed resistance mechanisms. | Patterson et al. [123] |
| Molecular marker techniques, multi-omics techniques | To better understand the mechanisms of invasive weeds. | Huang et al. [124] | |
| The WeedPedia platform | This review summarized the current status of weed genomics and introduced the International Weed Genomics Consortium (IWGC). | Montgomery et al. [125] | |
| QTL, Genome assembly | The author aimed at cruciferous weeds to study the early-growth of weeds and understand the origin of weeds. | Li et al. [126] | |
| Repetitive sequence analysis,Genome assembly | The genomic basis for the enhanced herbicide adaptability of polyploid Leptochloa chinensis was explored. | Chen et al. [127] | |
| Tomato | CRISPR/Cas9 | Promoting plant research and precision plant breeding. | Xia et al. [128] |
Zhang et al. [129] introduced the current status of genomic mating in poultry, which was a new method of animal breeding and still need continue to be combined with bioinformatics to handle large amounts of data in the future. Chicken(Gallus gallus domesticus) is the most widely farmed poultry. Li et al. [130] demonstrated that combining GWAS/SNPS and machine learning improved the accuracy of chicken genome prediction. Wang et al. [131] explored the field of RNA editing in chickens by establishing an RNA database of chickens combined with whole-genome analysis. In addition to these advances in genomic technologies, blight was also an important threat to chickens. Zhou et al. [132] introduced a genomics laboratory, GIP IL, which carried out genomic prediction and selection through GWAS and sequencing SNP panel. Excellent varieties resistant to Newcastle disease (ND) were bred. Smith [133] reviewed the application of genomics to animal defense against various avian viruses, such as ND, avian influenza, and poxvirus. Chen et al. [127] identified gene silencing by RNAi as a novel tool for mite control by analyzing core RNAi pathway genes in Dermanyssus gallinae. Feng et al. [134] used poultry in the southern United States as experimental materials to analyze E. coli through molecular markers and comparative genomics, which was helpful for the development of corresponding vaccines. In addition, in order to breed excellent varieties. Olaniyan et al. [135] reviewed methods to integrate community-based breeding programmes with genomics to facilitate breeding improvement in chickens. Wadood et al. [136] review the application of genomics to improved chicken reproduction, including techniques for epigenomics and precision gene editing. Long [137] investigated ways to improve the accuracy of predicting male fertility using genomic tools and identified markers such as the TSPY and PRM genes. Nematbakhsh et al. [138] designed a new breed of local village chicken with discrimination by genome-wide SNP. To prevent commercial fraud by selling colored broiler chickens as if they were local village chicken. High-quality databases are essential for genomics research. Barros et al. [139] used long-read technologies to identify new target genes and assemble a new turkey genome, providing a wealth of genomic data. Li et al. [140] presented for the first time a genomics database of helmeted guinea fowl and introduced its use. Li et al. [140] combined multiple sequencing techniques for de novo genome assembly and established a de novo pan-genome of 20 chickens to provide genomic data for breeding improvement of chickens.
Ducks(Anas) are also farmed in large numbers around the world. Che et al. [141] demonstrated the advantages of HiFi reads technology in identifying duck genomes by RNA-seq and identification of differentially expressed genes using healthy female mule ducks as experimental material. Guo et al. [142] explored the complex origin of domestic ducks by introgression analysis of whole genome sequences of mallards, maple leaf ducks, and domestic ducks. Chang et al. [143] used Chinese crested duck as experimental material, and through construction genome, comparative genomics analysis and SV, adaptive evolution and the origin of CC duck were further explored. Lin et al. [144] used Chaohu and Ji 'an Red ducks as experimental material to understand the differentiation process of ducks based on the analysis of population structure and candidate genes. To select for superior traits, Du et al. [145] combined genomics data with bioinformatics tools to investigate the molecular mechanisms by which ducks stop laying eggs, as well as changes in the liver that contribute to increased egg production. Zhang et al. [146] identified MC1R as the key gene regulating feather color through genotyping, SNP, and GWAS in a large number of ducks of different species. Yu et al. [147] investigated the mechanism of artificial selection by resequencing using Peking duck as experimental material.
Recent studies by various research teams have utilized genomic data and analyses to investigate the origin, introgression, and superior traits of different goose(Anser cygnoides) breeds, providing valuable insights and candidate genes for breeding improvements. Wen et al. [148] used the sequencing data of swan and greylag geese to understand the origin and introgression of Chinese geese through demographic analysis and candidate gene annotation analysis. Zhang et al. [149] performed a high-quality genome assembly of wild swan goose, providing a high-quality reference genome. Zhou et al. [150] targeted feather sac development in The Hungarian White Goose and generated high-quality genome-wide data that could help improve breeding. Liu et al. [151] identified FAP and GCG as candidate genes for high egg production by signal analysis based on SNP and qRT-PCR, which simultaneously contributed to the selection of Wulong geese with high egg production. Zhang et al. [152] conducted Linkage disequilibrium analysis on genome-wide data and identified several candidate genes, such as TGFBR3L, that were associated with superior traits in geese, which could help goose breed improvement.
Animal husbandry is important for the nutrient supply of the human diet, and the development of genomics is important for animal husbandry. Cows(bovis) can provide nutritious meat and milk. Guinan et al. [153] confirmed the role of GS on breeding improvement by analyzing genetic data from the National Cooperator Database and investigating multiple dairy cow breeds. Kertz et al. [154] theoretically reviewed the application of SNPS and other genomic methods to Improve Female Fertility, which still needs further practice in the future. Du et al. [155] reviewed the application of genomics in understanding the genetic mechanisms of cattle for main bovine species. Tian et al. [156] reviewed the role of GWAS, GP, and GS in breeding high-quality beef. Aiming at the quality and yield of dairy cattle, Zhao et al. [157] constructed the genome of predicted genes for cows with A2A2 casein genotype by CDHIT (v4.5.7) software, and analyzed their rumen fluids. The signature metabolite AA was found to play a key role in milk quality. Billa et al. [158] performed feed restriction by grouping experiments and extracted mirnas for microarray analyses and bioinformatic analyses, respectively, mirna was found to regulate milk yield by affecting mammary structure and lipid biosynthesis. Zavadilova et al. [159] used genome-wide association and genetic evaluation to improve disease resistance in dairy cows.
Pigs(Sus scrofa) are also one of the most important livestock. Yin et al. [160] provided valuable genomic data on Ningxiang pigs by performing Runs of homozygosity as well as genomic breed composition. Li et al. [161] performed gene introgression scan on Yunnan indigenous pigs and European pigs, through specific gene loci such as NAF1, The discovery of population gene flow in pigs helps to understand the genetic mechanisms behind traits and ultimately drives. Piorkowska et al. [162] review recent advances in pig genomics, including exploration of the origins of pig domestication, genes involved in lipogenesis, and candidate genes for litter size. Johansson et al. [163] compared two types of porcine kidney epithelial samples by short-read sequencing and found aneuploidies, which contributed to further understanding of livestock genomics.
In addition, the horse(Equus caballus) industry is also an important part of the livestock industry. Flack et al. [164] used Oxford Nanopore sequencing to construct a high-quality genome assembly for The Przewalski's horse breed, providing valuable genomic data. Littiere et al. [165] identified 53 candidate genes through a genome-wide association study and candidate gene analysis to understand the genetic map of horses of various breeds. Polak et al. [166] demonstrated the application of genomics in exploring inbreeding in rare horses by pedigree analysis of Sokolski horses and Sztumski horses using biological software Admixture. Frohlich et al. [167] conducted a study on Timor ponies (TP) to investigate the genetic relationship of TP through genotyping, SNP and ROH detection, which is helpful for breeding studies of TP.
Finally, Saleem et al. [168] comprehensively reviewed the major development of Nutrigenomics in animal husbandry, which has broad development prospects for improving animal yield and traits by exploring the relationship between genes and nutrition. A summary of the application of genomics in other animals is provided in Table 2.
Fishing is an important part of agriculture, with a rich diversity of fish species, but it also needs to be protected. To enrich fishery genomics resources, Alvarenga et al. [177] constructed 70 fish mitochondrial genomes through custom baits and target enrichment, which are molecular resources for fish management. Papa et al. [178] provided a high-quality population genomics resource by assembling the genome of the marine teleost tarakihi by combining short Illumina reads and long nanopore reads.
Bertram et al. [179] conducted SNP genotyping and bioinformatics analysis on snapper(Lutjanus erythropterus) from the coast of Western Australia, and investigated the population structure of snapper using population genomics. A more continuous genome assembly by long-read sequencing by Blommaert et al. [180] identified multiple structural variants, which is using genomics to facilitate fisheries management. Timm et al. [181] used community genomics to evaluate several oceanography models, including features such as spawning grounds, to help increase reproduction of Mexican Pink Shrimp(Penaeus duorarum) as well as to understand their evolutionary history. Ferrette et al. [182] performed de novo Genome Assembly and established a new reference genome for ssailfish(Istiophorus albicans), which is beneficial for understanding ssailfish speciation and may advance comparative genomic. Woodings et al. [183] used ddRADSeq and SNP to investigate the population structure of Eastern Rock Lobster to assess the harm of overfishing. Timm et al. [184] review the current issues to be addressed by Seascape genomics and its importance to maintaining a sustainable fishery. Oppen et al.[185] reviewed the current widespread use of genomics in marine conservation, demonstrating the need to use genomic monitoring and reference genomes to conserve marine diversity. Caccavo et al. [186] generated whole-genome libraries by small-scale sequencing of otoliths to provide genomic data for fisheries genomics. Casas et al. [187, 188] review the application of multiple emerging genomic technologies such as high throughput molecular technologies and RAD-seq in fisheries. Andersson et al. [189] reviewed methods of population genomics to promote sustainable fisheries, such as SNP-chip analysis and genome prediction, among others. Using golden perch as experimental material, Attard et al. [190] proposed that detection of ancestry of broodstock through genomics could promote the integration of genomics and fishery management. A summary of the application of genomics in other animals is provided in Table 2 .
| Subjects | Technology | Conclusion | References |
|---|---|---|---|
| GS, SNP, GWAS | Efficiency of genetic improvement to increase fertility in bulls. | Taylor et al. [169] | |
| Sheep,goat | Gene families clustering analysis, RNA sequencing | Understanding the taste of goat milk at the genomic level. | Zhang et al. [170] |
| GWAS, QTP, | To understand the genetic mechanisms of adaptation to extreme conditions in goats. | Li et al. [171] | |
| WGS data, phylogenetic analysis | To explore gene flow through Y chromosome variation in goats. | Nijman et al. [172] | |
| Pedigree data, Phenotypic data | Comparison of breeding value and genomic differences between Sanin and Alpine goats. | Negro et al. [173] | |
| SNPs, detecting InDels | A goat genome-wide data platform, the VarGoats project, was introduced. | Denoyelle et al. [174] | |
| single-nuclei ATAC-seq | This paper summarizes the achievements of single-cell omics research and analyzes the development prospects of this technology. | Lyons et al. [175] | |
| chicken | CRISPR/Cas9, base editing | To generate valuable poultry genome editing models. | Park et al. [176] |
Forestry plays an important role both in protecting diversity and in the economy [191]. But forestry breeding has a longer cycle than crop breeding, so the application of genomics is even more important. Genomic technologies are widely used in forestry [192]. Cao et al. [191] reviewed the application of CRISPR-Cas in forest trees, which has been used in fruit trees but still needs technological breakthroughs to improve efficiency and quality.
Mostert et al. [193] studies on the domestication of wood fiber crops by SNP genotyping can provide insights into evolutionary history and population structure. Anders et al. [194] reviewed the role of Cas-DNA-free technology and multiplex editing in tree breeding improvement. Ahmar et al. [195] reviewed the role of genotyping and TILLING screening in GAB for advancing forest tree breeding. In addition, many scientists have provided valuable forest genomics resources. Burley et al. [196] studied two tree species Jacaranda copaia and Handroanthus. By combining Illumina short-read and Chicago libraries data and using whole genome alignment technology, we found gene duplication between them. Niu et al. [197] reviewed recent advances in poplar genomics, from haploid sequencing to the realization of telomere to telomere (T2T), through length long sequencing technology, helping to advance breeding. Tumas et al. [198] used SNP technology to construct genetic maps of Picea sitchensis and Picea glauca, which is helpful for the development of genotyping chips and sustainable management of forest tree species. Capblancq et al. [199] revealed the genetic history of oriental beech and provided a quality genomics model by studying its whole genome data in the Caucasus region. Luo et al. [200] reviewed Dipterocarpaceae genomic data, combined with comparative genomics and phylogenetic analyses, to help select superior Dipterocarpaceae tree species using GS. Han et al. [201] established and analyzed the genome structure of Quercus variabilis by PacBio HiFi sequencing, which is conducive to comparative genomics studies. Matallana-Ramirez et al. [202] used Loblolly Pine as an example to illustrate the important role of genomics in managing forest ecosystems by discussing the development status of phenotypical genotype studies. Shirasawa et al. [203] analyzed four conifer species by HiFi long-read sequencing and found associations among them, helping to speed up the breeding process through genomic data. Finally, the impact of climate change on forests is also significant. Based on high-throughput phenotyping technology, Feng et al. [204] proposed combining landscape genomics and evolutionary genomics to cultivate trees that intelligently respond to climate change. Sharma et al. [192]reviewed the application of genotyping, QTL, and genome prediction in tree breeding. Abiri et al. [205] reviewed the effects of rainfall and other environmental stresses on forest trees, as well as the analysis of tolerance mechanisms of forest trees.
In the past five years, the application of genomics in agriculture has achieved remarkable results, especially in improving crop yield, quality, stress tolerance and adaptability, and promoting sustainable development of animal husbandry and fisheries. With the help of GAB technology, scientists can accurately improve crop and animal varieties to effectively meet the dual challenges of global food demand growth and environmental protection.
In the future, the application of genomics in agriculture will be more promising. With the continuous progress of technology and the emergence of new tools, genomics is expected to further enhance agricultural productivity, improve the quality and safety of agricultural products, and provide strong support for the sustainable development of agriculture and the environment. The integration of genomics with emerging technologies such as big data, artificial intelligence and biotechnology will inject new impetus into agricultural innovation and transformation. Continued investment in genomics research and its application in agriculture is important to meet global food demand, ensure food security and protect the environment. In addition, the potential of genomics to address climate change, such as breeding drought and heat tolerant crop varieties and protecting biodiversity through optimized population management and habitat restoration, deserves further exploration. The future of agriculture will rely on genomics to build more resilient, productive, and sustainable food systems for present and future generations.
 Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. Agricultural Science and Food Processing
ISSN: 3066-1579 (Online) | ISSN: 3066-1560 (Print)
Email: xucm@btbu.edu.cn

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/iece/