IECE Transactions on Sensing, Communication, and Control
ISSN: 3065-7431 (Online) | ISSN: 3065-7423 (Print)
Email: [email protected]


The high rates of morbidity and mortality caused by brain tumors make them a significant public health concern on a global scale. Brain tumors are either benign or malignant; the latter group is characterized by aggressive activity and fast growth, which frequently leads to a bad prognosis for patients. To choose the most appropriate treatment, such as surgery, chemotherapy, or radiation therapy, a prompt and precise diagnosis is essential. Nevertheless, identifying brain tumors manually using MRI scans is an incredibly difficult and expert-level undertaking. Diagnosis becomes more complicated due to the heterogeneity of malignancies and the numerous distinctions between normal brain components and cancers. Poor patient outcomes may result from less-than-ideal treatment options caused by incorrect or delayed diagnoses. Automatic techniques that can aid doctors in classifying brain tumors have, thus, been increasingly popular in recent years [1].
Magnetic Resonance Imaging (MRI) is the standard imaging modality used for diagnosing brain tumors, as it provides detailed information about soft tissues and is non-invasive. While MRI scans offer high-resolution images, the manual review process is both time-consuming and prone to variability among radiologists [2]. Given the large volume of scans produced in modern healthcare settings, there is an urgent need for computational tools that can reliably classify brain tumors and reduce the diagnostic burden on healthcare professionals [3, 4]. The advancement of machine learning, particularly deep learning, has opened new possibilities for the development of automated systems capable of performing accurate and efficient image analysis [5]. These techniques have proven to be especially effective in complex tasks such as medical image classification and segmentation, due to their ability to learn and generalize from large datasets.
In recent years, deep learning algorithms, specifically Convolutional Neural Networks (CNNs), have emerged as the leading approach for image classification tasks. CNNs have revolutionized the field of computer vision by their capacity to automatically extract hierarchical features from input images without the need for extensive manual feature engineering. This has made them particularly suitable for medical imaging tasks, where the inherent complexity and variability of the images pose significant challenges for traditional machine learning methods [6, 7]. CNNs are composed of multiple layers, including convolutional, pooling, and fully connected layers, which progressively learn features ranging from simple edge detection to complex patterns specific to the task at hand. This enables CNNs to model the intricate relationships within image data, thereby significantly enhancing their classification accuracy [8, 9, 10, 11].
Use of convolutional neural networks (CNNs) for MRI-based tumor classification is the primary emphasis of this research [12, 13]. We explored two CNN architectures with different configurations to assess their efficiency in correctly diagnosing brain cancers as either tumor-positive or tumor-negative [14]. The first architecture utilizes single convolutional layers, while the second builds upon this by introducing double convolutional layers [15]. The rationale behind this exploration is to determine whether deeper network architectures, involving more convolutional operations, can provide a tangible improvement in classification performance [16]. Deep networks have the potential to capture more complex features from the images, leading to better generalization [17]. However, they also come with the risk of overfitting, especially when trained on smaller datasets, which necessitates careful evaluation and comparison between models [18, 19, 20].
In the first model, we implemented a relatively shallow CNN architecture, consisting of a single convolutional layer followed by pooling and fully connected layers. This simple yet effective structure achieved an outstanding accuracy of 99.6% in classifying brain tumors. This result underscores the power of even a single-layered CNN in identifying critical features within MRI scans [21, 22]. However, as deep learning models excel at acquiring increasingly intricate patterns as their training data sets become more extensive, we extended our study by designing a second CNN model with double convolutional layers. This architecture aims to further explore the performance benefits of deeper feature extraction, where consecutive convolutional layers may allow the network to learn more abstract and meaningful representations of tumor characteristics.
Our investigation provides a thorough comparative analysis between these two CNN-based me,thods in terms of classification accuracy, computational efficiency, and robustness to overfitting. The performance metrics of these models are evaluated on a dataset of brain MRI images, containing both tumor and non-tumor data [26, 27]. By assessing the outcomes of single-layer versus double-layer CNNs, we aim to provide valuable insights into the trade-offs between model simplicity and performance. The primary contributions of this research are twofold: first, demonstrating the efficacy of CNNs in brain tumor classification with high accuracy; and second, offering a detailed exploration of how network depth influences classification performance in this context.
The results of this investigation have significant implications for the design and deployment of automated diagnostic tools in clinical practice. Automated brain tumor classification systems have the potential to augment the diagnostic capabilities of radiologists, reducing the time required for manual assessments and improving diagnostic consistency. Moreover, these systems could assist in early-stage tumor detection [23], where subtle abnormalities may be easily overlooked by the human eye. By enhancing the speed and accuracy of diagnosis, deep learning-based systems can contribute significantly to bettering patient care and results in brain tumor cases [24, 25].
In the upcoming sections, we will discuss the dataset and preprocessing methods, the architectures of the CNN models employed, and the results of our comparative analysis. Finally, we go over some of the possible next steps for study in this area and the consequences of our conclusion.
Gómez-Guzmán et al. [19] tested seven models of deep convolutional neural networks using the Msoud dataset for brain tumor MRI. Their preprocessing involved resizing, labeling, and data augmentation. The InceptionV3 model achieved the highest avg. accuracy of 97.12%, followed by ResNet50 and InceptionResNetV2 with 96.97% and 96.78%, respectively. A notable limitation of their work is the reliance on a single dataset, which may hinder generalization to other data sources. Reyes et al. [1] worked with the Figshare dataset of 3064 T1-weighted MRI images and the Kaggle dataset of 3264 MRI slices. Using neural networks for Feature extraction and Classification, they achieved a best accuracy of 98.7% using models like ResNet50 and Vision Transformers.
| Ref. No. | Author | Dataset | Method | Results | Advancement/ Future Directions |
|---|---|---|---|---|---|
| 2023 [19] | Guzmán et. al. | Brain Tumor MRI dataset Msoud | Deep CNN, Preprocessing with resizing, labeling, data augmentation | GCNN 81.05%,InceptionV3 97.12%, ResNet50 96.97%, ResNetV2 96.78% | Diverse dataset, lower computation cost, Integration of multimodal imaging data |
| 2024 [1] | Reyesa et al. | Figshare dataset, Kaggle dataset: 3264 MRI | Hybrid methods combining NNs, ML methods | 98.7% accuracy in MobileNet and EfficientNet | extended analysis, additional NNs for classification |
| 2024 [2] | Srinivasan et al. | Custom dataset | Deep CNN, Grid search, ROC curve | 1st CNN 99.53%, 2nd CNN 93.81%, 3rd CNN 98.56% | Integration of real-time data for dynamic classification |
| 2023 [3] | Saeedi et al. | 3264 T1-weighted MRI images | 2D CNN, auto encoder, 6MLs, ROC curve | 2D-CNN 96.47%, Roc 0.99, KNN 86% | Hybrid CNN with MLs. applications in clinical settings |
| 2018 [29] | Seetha et al. | Radiopaedia Brats 2015 | CNN, Fuzzy-C, LIPC for voxel, Cellular Automata. | CNN achieved 97.5% accuracy | Accuracy Improved, Reduce compute time |
| 2024 [18] | Zahoor et al. | Kaggle, Br35H, and Figshare repositories | novel deep residual and regional CNNs | Res-BRNet 98.22%, F1-score 0.9641 | More advanced augmentation techniques, with limitations in misclassification val |
| 2022 [25] | Younis et al. | BRATS 2013 Dataset, World Brain Atlas | CNN using VGG16 | CNN accuracy 96% | Diversity for brain region approximation |
| 2023 [16] | Atasever et al. | ImageNet, MICCAI 2012 PROMISE12, COVID-chest X-ray-dataset | Capsule Network-based framework | Hybrid model with pr-trained having 92.68% | AI diagnosing gastrointestinal diseases using wireless-capsule endoscopy |
| 2023 [17] | Aktera et al. | 4 Datasets a, b, c, d | CNN with U-Net-based model | 98.7% for merged dataset | exploring additional datasets, and further clinical validation |
However, their work was limited by the lack of diversity in the datasets, which restricts the applicability of the findings to broader cases. Srinivasan et al. [2] used a custom dataset with deep CNNs for classification, hyperparameter tuning via grid search, and fivefold cross-validation. Their models achieved high detection and classification accuracies, including a peak of 99.53%. A significant drawback of this study is the absence of testing on varied datasets, limiting its robustness in different scenarios. Saeedi et al. [3] employed a dataset of 3264 T1-weighted MRI images containing glioma, meningioma, pituitary, and healthy cases. They used 2D CNNs and convolutional autoencoders for tumor classification, achieving accuracies of 96.47% and 95.63%, respectively. One challenge in their work was the relatively low performance of traditional ML methods compared to deep learning approaches.
Seetha et al. [29] utilized the Radiopaedia and Brats 2015 datasets. They applied CNNs for classification alongside fuzzy C-means for segmentation, achieving a classification accuracy of 97.5%. A limitation of their study is the significant computation time required for classification, which may hinder real-time applications. Zahoor et al. [18] proposed a novel Res-BRNet architecture combining residual and regional CNNs for brain tumor classification. Using datasets like Kaggle and Figshare, they achieved an accuracy of 98.22% and an F1-score of 0.9641. A weakness in their approach is the persistence of residual misclassification rates, which could impact diagnostic reliability. Younis et al. [13] had used datasets like BRATS 2013 and World Brain Atlas to employ CNNs, particularly VGG16, for tumor identification, achieving 96% accuracy. Future work involves using diverse modalities and enhancing ensemble methods for precision. Atasever et al. [16] used datasets such as MICCAI 2012 and BraTS for a capsule network framework. Their hybrid model achieved a 92.68% accuracy in disease diagnosis. However, the model’s limitations include relatively lower accuracy compared to state-of-the-art methods and inadequate generalization across multiple datasets. Akter et al. [17] worked with individual and merged datasets to propose CNN-based classification and U-Net-based segmentation models. Achieving accuracies of 98.7% and 98.8%, the study’s primary limitation lies in the insufficient exploration of model efficiency and lack of thorough clinical validation. The Table 1 illustrates a comparative analysis of recent research pertaining to the classifications and segmentation of brain tumors.
There are many methods present through which we can perform brain tumor detection; to name some of them, they are RNN [30], LSTM [31, 32, 33], deep learning [28], and transfer learning [26]. Our model uses a convolutional neural network to perform the classification task effectively. The structural flow followed by the model is as shown in Figure 1.
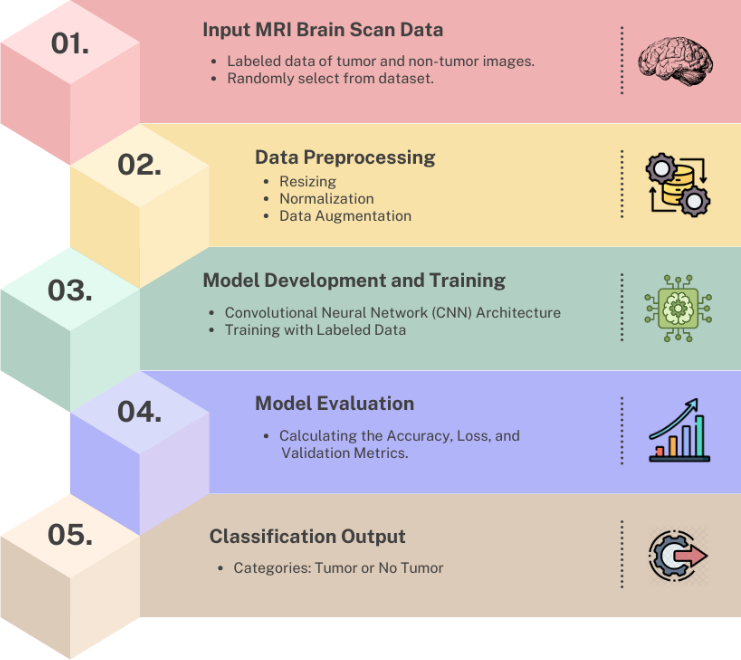
To ensure uniformity and enhance model performance while preventing overfitting, MRI images underwent preprocessing involving rescaling, data augmentation, and dataset splitting. The images were converted to grayscale, resized to 150×150 pixels, and normalized to a [0,1] range, which improved computational efficiency and ensured consistent weight updates during training. Data augmentation, essential due to the modest dataset size of 253 images, artificially increased diversity by applying random rotations, horizontal and vertical flips, and zooming, dynamically generating unique variations in each epoch to reduce overfitting and enhance generalization. Sections for training (70%), validation (15%), and testing (15%) were created from the dataset., using stratified splitting to maintain balanced class distributions. Slight class imbalance, with 153 tumor and 98 non-tumor images, was addressed through oversampling during data augmentation, ensuring equalized training set sizes.
The CNN architecture, as shown in Figure 2, was meticulously designed to handle the complexity of MRI images while being computationally efficient. The model leverages the hierarchical feature extraction capability of convolutional layers, progressively learning to detect features like edges, textures, and higher-level tumor structures.
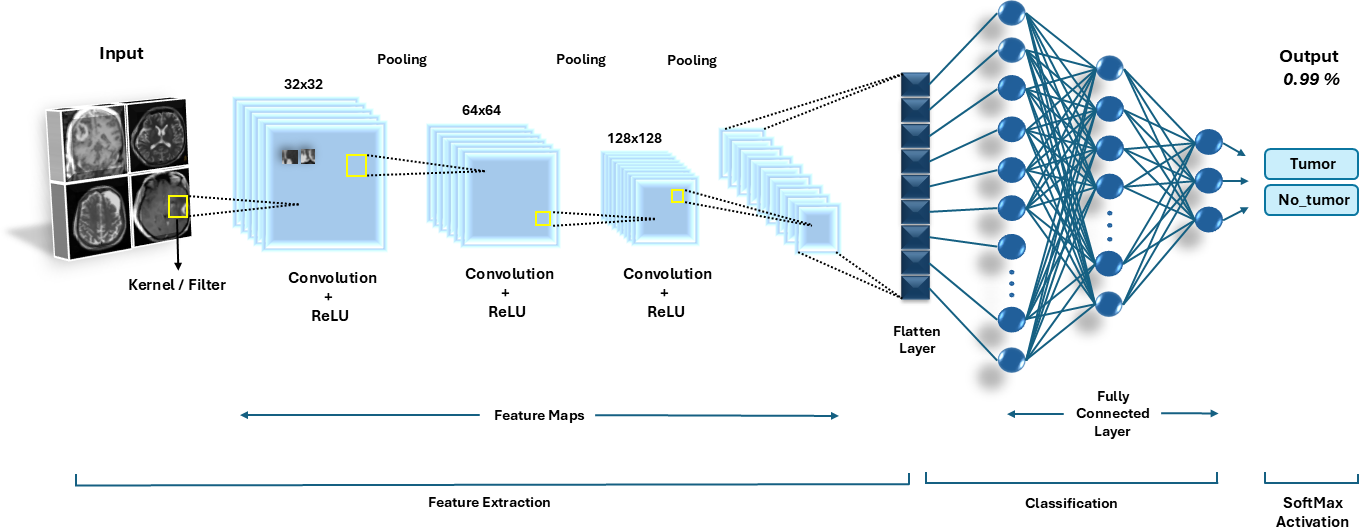
The model, as depicted by Table 2, consists of three convolutional blocks with filters starting at 32 and doubling at each subsequent block (32, 64, 128), where convolution operations detect spatial features applied after each convolutional block to downsample feature maps, reducing computational load while retaining critical features. The output of the final convolutional block is flattened into a 1D vector to prepare the data for dense layers. Activated by ReLU, a fully linked dense layer consisting of 512 neurons produces a high degree representation of the features that have been retrieved. To prevent overfitting, a dropout layer with a 50% rate is employed after the dense layer, randomly deactivating neurons during training.
A sigmoid-activated single neuron makes up the output layer, producing probabilities between 0 and 1, indicating the likelihood of the input image containing a tumor.
| Layer Type | Filters/Units | Output Shape | Parameters |
|---|---|---|---|
| Convolutional Layer | 32 filters | (150, 150, 32) | 896 |
| Max Pooling | - | (75, 75, 32) | 0 |
| Convolutional Layer | 64 filters | (75, 75, 64) | 18496 |
| Max Pooling | - | (37, 37, 64) | 0 |
| Convolutional Layer | 128 filters | (37, 37, 128) | 73856 |
| Max Pooling | - | (18, 18, 128) | 0 |
| Flatten | - | (41472) | 0 |
| Dense Layer | 512 neurons | (512) | 21235200 |
| Dropout | - | (512) | 0 |
| Output Layer | 1 neuron | (1) | 513 |
The multilayer CNN architecture, as depicted by Table 3, begins with an input layer of size 224x224x3 to process RGB images. To begin extracting basic spatial characteristics, the first convolutional layer uses 64 filters with a 3x3 kernel size and a ReLU activation function. The spatial dimensions are subsequently reduced using a max pooling layer with a 2x2 pool size. The second convolutional layer increases the filters to 128 with a kernel size of 3x3, followed by another ReLU activation for deeper feature extraction and a max pooling layer with a pool size of 2x2. The third convolutional layer further enhances the network’s capacity with 256 filters, a kernel size of 3x3, and ReLU activation, followed by max pooling to reduce the feature map size.
The output of the convolutional blocks is flattened and passed to the first fully connected layer with 512 neurons and ReLU activation to combine learned features. A second fully connected layer with 256 neurons and ReLU activation further refines these features. Finally, the output layer, with SoftMax activation, provides the classification probabilities for the target classes.
| Layer Type | Filters/Units | Output Shape | Parameters |
|---|---|---|---|
| Convolutional Layer | 32 filters | (150, 150, 32) | 896 |
| Max Pooling | - | (75, 75, 32) | 0 |
| Convolutional Layer | 64 filters | (75, 75, 64) | 18,496 |
| Max Pooling | - | (37, 37, 64) | 0 |
| Convolutional Layer | 128 filters | (37, 37, 128) | 73,856 |
| Max Pooling | - | (18, 18, 128) | 0 |
| Convolutional Layer | 256 filters | (18, 18, 128) | 73856 |
| Max Pooling | - | (9, 9, 256) | 0 |
| Flatten | - | (207,936) | 0 |
| Dense Layer | 1024 neurons | (1024) | 214,994,240 |
| Dropout | - | (1024) | 0 |
| Dense Layer | 512 neurons | (512) | 524,800 |
| Dropout | - | (512) | 0 |
| Output Layer | 1 neuron | (1) | 513 |
| Total Parameters | - | - | 214,888,496 |
The model’s final steps included optimization, loss computation, and training configuration. The Adam optimizer was employed for its adaptive learning rate and computational efficiency, updating weights based on the first and second moments of gradients for faster convergence. Binary Cross-Entropy loss, suited for binary classification, was used to quantify the error between predicted probabilities and actual labels, calculated as in equation (1):
A batch size of 32 was chosen to strike a balance between computational efficiency and robust gradient estimates. The model was trained for 20 epochs, providing sufficient learning time without risking overfitting, ensuring it effectively captured patterns for binary classification tasks.
Below given Table 4, examines the key aspects such as network depth, parameter count, generalization capabilities, accuracy, and training time.
| Aspects | Std. CNN | Multi-Layer CNN |
|---|---|---|
| Depth | Shallower network (3 convolutional layers) | Deeper network (e.g., 5+ layers) |
| Parameters | Lower parameter count | Higher parameter count |
| Generalization | Suitable for smaller Datasets | Better suited for larger datasets |
| Accuracy | Achieved: 99.60% | Achieved: 86% |
| Training Time | Faster | Slower due to increased complexity |
This section encompasses the dataset utilized in this research. Performance is evaluated through a range of indicators, specifically the evaluation metrics. The following section presents the results through graphical visualization and precisely classified images.
For this research, we employed a publicly available MRI brain scan dataset sourced from Kaggle. This dataset comprises a total of 253 images, with 153 labeled as having brain tumors and 98 as non-tumor cases. The dataset provides a balanced representation of both tumor and non-tumor images, facilitating comprehensive analysis and model training. Utilizing this data, our study aims to improve classification accuracy and contribute to the advancement of automated brain tumor detection methodologies in clinical settings. Images were resized to 150×150 pixels for uniformity. The dataset was split into training (70%), validation (15%), and test (15%) subsets, maintaining class balance in each set.
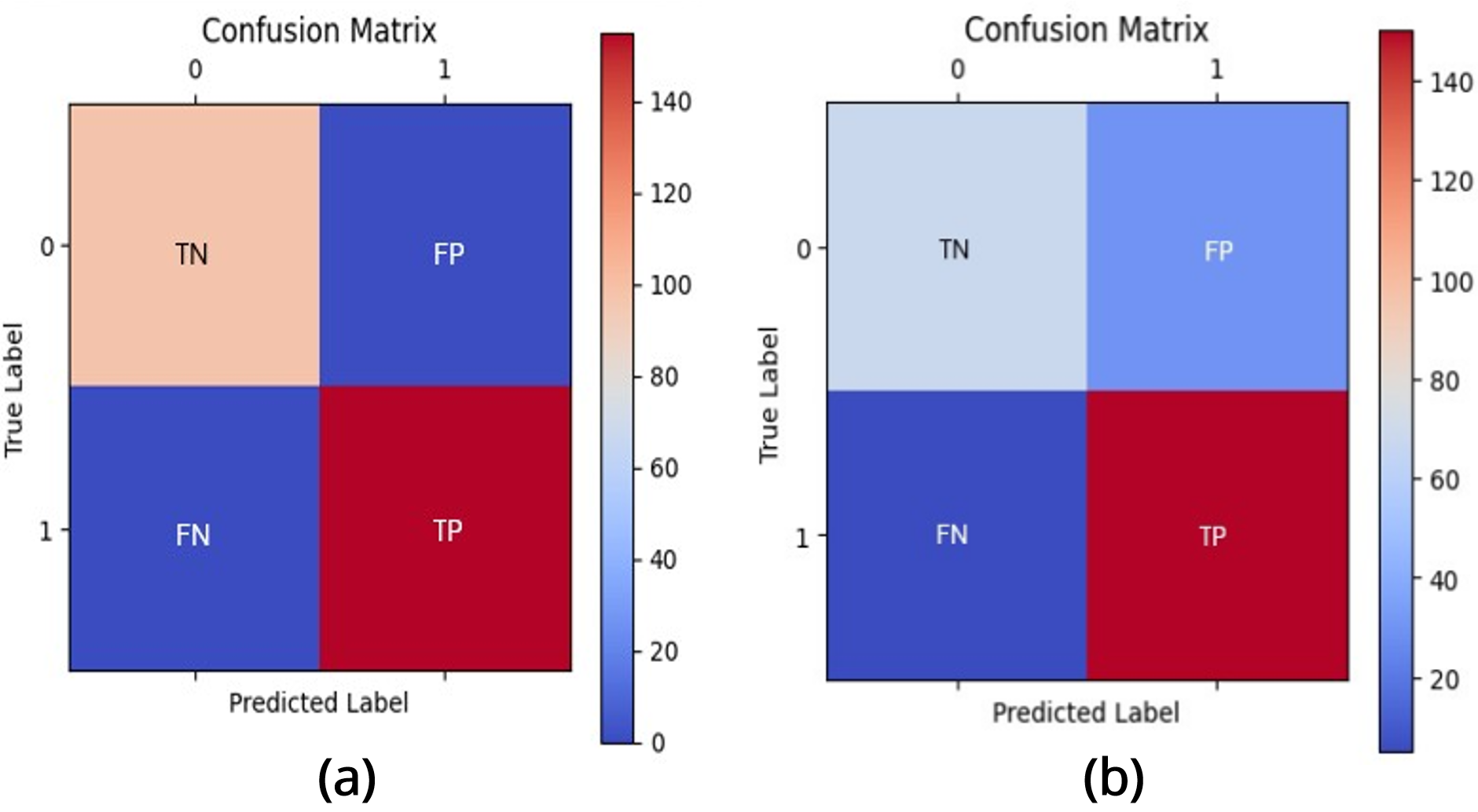
To evaluate the performance of the brain tumor classification model, we utilized various classification metrics, including accuracy, precision, recall, and F1 score. These metrics were calculated using the confusion matrix, which provides detailed insights into the model’s predictions.
Accuracy works by measuring the overall correctness of the model by calculating the ratio of correctly predicted instances to the total number of instances. In our project, accuracy indicates how often the model correctly classifies brain MRI images into tumor and non-tumor categories. Calculated using Equation (2):
Precision measures the proportion of correctly predicted positive observations out of all predicted positives. Precision is crucial for minimizing false positives, which ensures that non-tumor cases are not incorrectly classified as tumor cases. This is particularly significant in medical diagnostics to prevent unnecessary stress for patients. Calculated using Equation (3):
Recall also known as sensitivity, calculates the proportion of correctly predicted positive observations out of all actual positives. In our project, recall is vital for identifying as many tumor cases as possible, ensuring that critical cases are not overlooked. High recall ensures that the model captures most of the positive cases, which is crucial in medical applications. Calculated using Equation (4):
F1 Score is the harmonic mean of precision and recall. It provides a balanced measure, especially when there is an uneven class distribution. The F1 score ensures that both precision and recall are balanced, which is particularly important when dealing with medical datasets where both metrics are critical. Calculated using Equation (5):
In the confusion matrix as shown in Figure 3, the True Negatives (TN), located in the top-left (00) cell, are cases where the model correctly predicts the negative class, such as "No Tumor" when the actual label is also "No Tumor." The False Positives (FP), in the top-right (01) cell, occur when the model incorrectly predicts the positive class, such as classifying "No Tumor" as "Tumor," which can lead to over-diagnosis. Conversely, the False Negatives (FN), found in the bottom-left (10) cell, represent cases where the model incorrectly predicts the negative class, such as failing to detect an actual tumor by predicting "No Tumor."
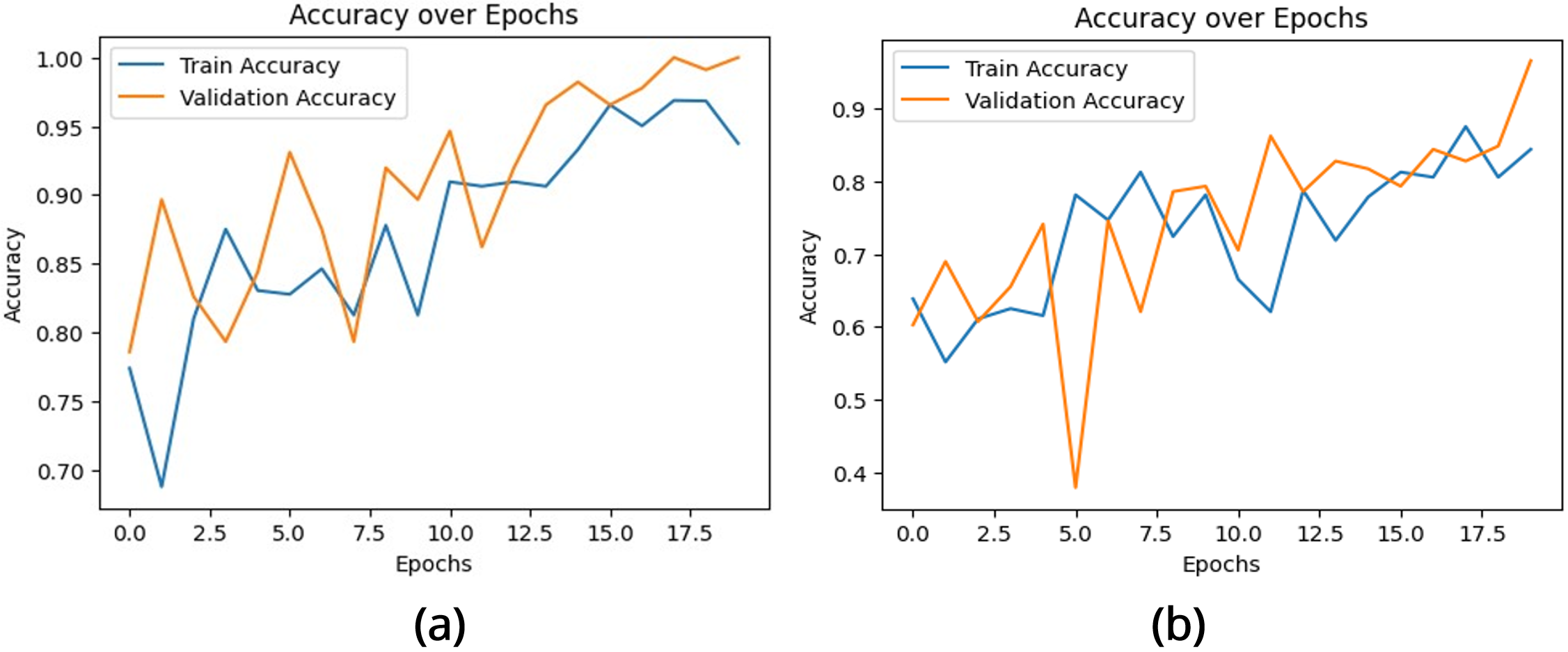
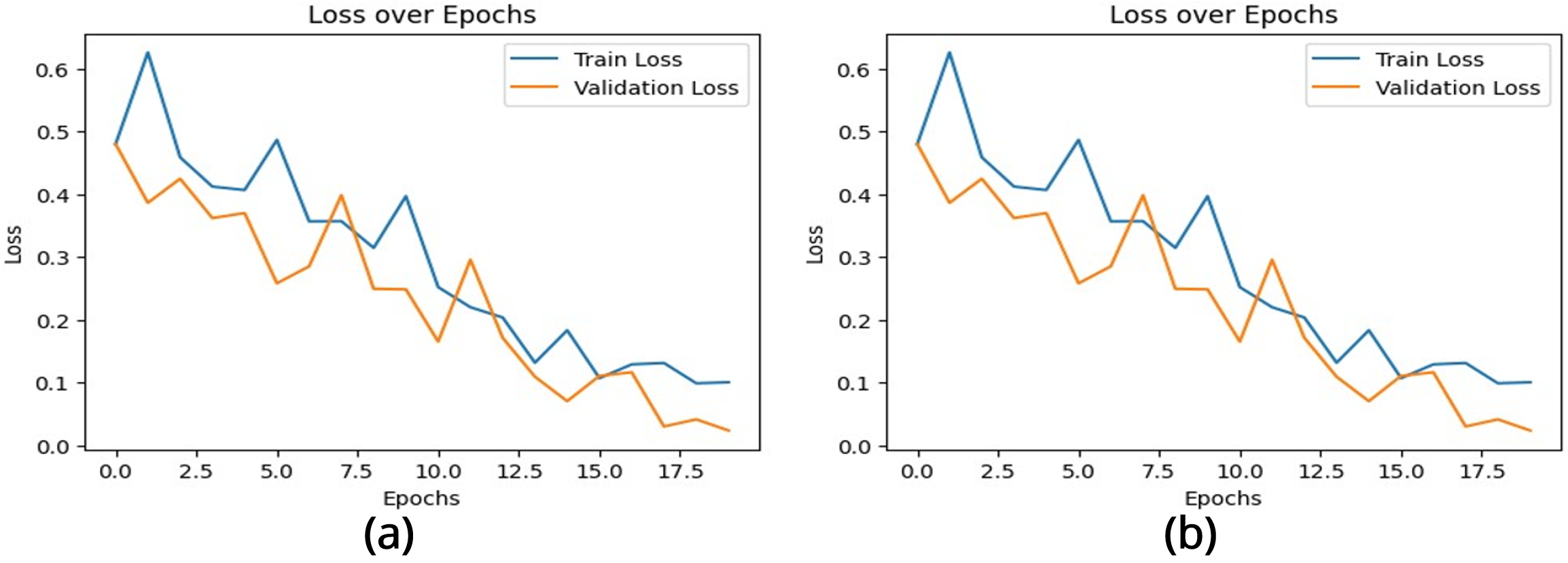
The performance of the brain tumor classification model was evaluated using the training and validation accuracy and loss curves, as shown in Figures 4 and 5. These metrics provide critical insights into the learning behavior and generalization capability of two CNN architectures over 20 epochs.
| Evaluation Metric | Score |
|---|---|
| Accuracy | 0.996 |
| Precision | 0.9936 |
| Recall | 1 |
| F1-Score | 0.9968 |
The accuracy curve Figure 4 (a) illustrates the proportion of correctly classified instances for both training and validation datasets. Initially, the model demonstrates moderate accuracy, reflecting its early stage of feature learning. Over successive epochs, the training accuracy exhibits a steady increase, converging near 1.0, which indicates effective learning of distinguishing features. The validation accuracy closely tracks the training accuracy, showing minor fluctuations due to dataset variability. By the final epochs, the near alignment of both curves indicates minimal overfitting and strong generalization to unseen data.
In contrast, the accuracy graph Figure 4 (b) of the multilayer exhibits’ significant fluctuations, particularly in the early epochs. While the validation accuracy eventually surpasses the training accuracy, this sharp difference indicates potential underfitting during the training phase. Despite the fluctuations, the final validation accuracy shows a marked improvement compared to the initial epochs, indicating that the model is learning, albeit with instability.
The loss curve Figure 5 (a) represents the error between predicted and actual outputs for both datasets. High initial losses decrease significantly during training, demonstrating the model’s optimization and convergence. The training loss exhibits a smooth decline, while the validation loss follows a similar trend with minor variability. By the final epochs, both losses stabilize at low values, indicating that the model has achieved effective feature extraction and prediction accuracy.
The multilayered CNN model’s loss graph Figure 5 (b) is characterized by notable fluctuations, particularly in the validation loss during the early and middle epochs. Although the training loss decreases steadily, the validation loss exhibits instability before a sharp drop in the later epochs. This behavior suggests potential noise in the dataset, overfitting during some phases of training, or sensitivity to hyperparameters. The sharp decline in loss at the end indicates eventual convergence, but the instability might hinder the model’s reliability.
These plots have collectively demonstrated that the CNN-based model exhibits a well-balanced learning process. The steady increase in accuracy, coupled with the consistent decrease in loss, highlights the model’s capacity to learn meaningful features for the classification of brain MRI images [34]. Moreover, the convergence of both training and validation metrics at similar levels indicates that the model is both reliable and robust, making it suitable for deployment in medical imaging applications where accurate and consistent predictions are critical.
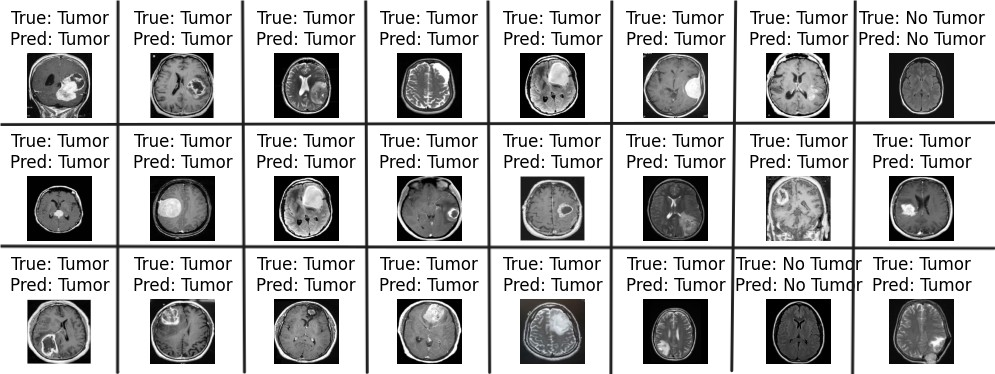
Firstly, this study proposed a standard convolutional neural network for the detection of brain tumors, which are very crucial diseases in the medical domain. The features used for the classification were extracted RGB color features, which quantify the relevance of a word to a document. The second phase method is applied using a multilayered CNN model, and various metrics like accuracy and precision are calculated to evaluate the performance of the classifiers. These results indicate that the standard CNN model generates better results compared to the multilayered CNN model, as mentioned in Table 5. If you look at Table 6, you can see that our model outperforms the others based on the comparison of performance metrics with previously published studies. Figure 6 demonstrates the accurate classification of brain tumor images based on our proposed model.
In order to enhance brain cancer care, it is necessary to understand and integrate the whole scope of cancer experiences. However, due to the unstructured nature of medical records, large-scale data integration using classical methods is very resource expensive. This void is being filled by DL methods, which have already transformed the gathering, processing, and understanding of often-gathered unstructured health data. This study proves CNNs can detect brain tumors from MRIs. The standard CNN had 99.60% accuracy for small datasets and faster training. Multilayer CNN had an accuracy of 86.17% but needed longer training and showed moderate overfitting, making it better for larger datasets but inefficient for smaller datasets. Stability and consistency make the first model better for brain tumor identification. The second model’s sophisticated architecture should improve performance, but it needs more optimization to stabilize training and validation metrics.
The model’s future development could include extending it to handle 3D MRI datasets for improved diagnosis, incorporating transfer learning techniques for limited datasets, and incorporating Grad-CAM for interpretability. Moreover, integrating advanced data augmentation techniques and hybrid models combining CNNs with transformers could enhance accuracy. Expanding dataset diversity and evaluating the model’s robustness across different imaging modalities and sensitivity across noisy data could also be valuable directions.
IECE Transactions on Sensing, Communication, and Control
ISSN: 3065-7431 (Online) | ISSN: 3065-7423 (Print)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/iece/