IECE Transactions on Emerging Topics in Artificial Intelligence
ISSN: 3066-1676 (Online) | ISSN: 3066-1668 (Print)
Email: [email protected]


Public health surveillance plays a vital role in detecting emerging health threats, monitoring disease outbreaks, and guiding timely interventions. Traditional surveillance systems primarily rely on structured data sources, such as hospital records, laboratory test results, and government health reports. While these methods provide critical insights, they often suffer from delays in data collection, processing, and reporting. Studies indicate that conventional surveillance systems can lag by days or even weeks in detecting outbreaks, limiting their effectiveness in real-time decision-making [1]. Furthermore, structured datasets alone may not capture early indicators of disease spread, especially in rapidly evolving epidemiological scenarios.
With the rise of digital platforms, vast amounts of unstructured data [2] are continuously generated from sources such as electronic health records (EHRs), social media platforms, online news articles, scientific literature, and patient forums like HealthBoards and MedHelp. Reports suggest that over 80% of healthcare data is unstructured, making it a valuable yet underutilized resource for disease monitoring. Social media platforms such as Twitter (now X) and Reddit have been shown to provide early warning signals for emerging diseases, as users often share symptoms and local outbreaks before official reports are published. However, extracting meaningful insights from such unstructured textual data presents significant challenges due to the volume, variability, and linguistic complexity of the information.
Recent advancements in Natural Language Processing (NLP) and Artificial Intelligence (AI) have revolutionized the ability to analyze unstructured text at scale. Transformer-based language models, such as BERT and GPT, enable accurate text classification, named entity recognition (NER), sentiment analysis, and topic modeling, facilitating automated public health intelligence. By leveraging NLP and AI-driven methodologies, public health agencies can detect early warning signals, identify emerging disease clusters, and track misinformation related to health crises. For instance, studies have demonstrated the effectiveness of NLP-based models in tracking influenza and COVID-19 trends using social media posts and online news [3, 4].
This research introduces an AI-driven NLP framework for automating disease surveillance using unstructured data. The proposed system integrates advanced NLP techniques to extract epidemiological insights, perform anomaly detection, and conduct geospatial trend analysis. The framework is designed to analyze multilingual data streams in real time, enhancing the responsiveness of public health authorities. Potential applications include monitoring respiratory illnesses (e.g., influenza, COVID-19), vector-borne diseases (e.g., dengue, Zika virus), and foodborne outbreaks.
By leveraging real-time unstructured data processing, the proposed approach has the potential to transform public health intelligence, improving outbreak detection, response times, and overall disease monitoring capabilities.
It proposes a modular NLP-AI framework capable of ingesting and analyzing large-scale unstructured data from social media, EHRs, news, and forums to extract early signals of disease outbreaks.
The system integrates transformer-based models (BERT, GPT, BioBERT) for classification, sentiment analysis, and named entity recognition tailored to the public health domain.
It incorporates real-time anomaly detection, geospatial trend mapping, and multilingual support using cross-lingual transfer learning to enable global applicability.
The framework includes interpretable AI components (LIME, SHAP), federated learning with differential privacy, and adaptive learning mechanisms to ensure continuous performance and ethical operation.
The system's scalability and effectiveness are demonstrated through real-world case studies and simulations of pandemic-scale health data streams.
The integration of Natural Language Processing (NLP) and Artificial Intelligence (AI) in public health surveillance has gained significant attention in recent years. Existing research has explored various methodologies for extracting epidemiological insights from unstructured data sources, such as electronic health records (EHRs), social media, news articles, and online health forums. This section reviews relevant literature on AI-driven disease surveillance, NLP techniques for health intelligence, and applications of deep learning models in epidemiology.
Traditional disease surveillance methods, which rely on structured data from hospitals and laboratories, often suffer from delayed reporting and limited coverage [1]. AI-driven surveillance systems aim to overcome these limitations by leveraging machine learning and NLP techniques to analyze unstructured data sources in real time. Studies have demonstrated that social media platforms, such as Twitter and Reddit, provide early warning signals for emerging disease outbreaks. For example, the HealthMap project utilizes automated data mining to extract epidemiological insights from news reports and online content, contributing to real-time disease tracking [5].
Recent advancements in deep learning have further enhanced the ability to process and interpret unstructured health data. Transformer-based models, such as BERT and GPT, have been used for real-time disease classification and outbreak prediction [6]. Additionally, hybrid models combining NLP with geospatial analysis have been developed to map the spread of infectious diseases and identify high-risk regions [7, 18]. However, while AI-driven approaches significantly improve surveillance efficiency, ethical concerns arise regarding the collection and analysis of publicly shared health data. Studies highlight the need for privacy-preserving techniques and data anonymization to ensure ethical compliance in AI-driven public health applications [16].
NLP has emerged as a powerful tool for extracting and analyzing health-related information from large-scale text data. Named Entity Recognition (NER) enables the identification of disease names, symptoms, and locations in textual data, while sentiment analysis helps assess public perception and concerns regarding health crises [9]. Topic modeling techniques, such as Latent Dirichlet Allocation (LDA) and dynamic topic modeling, have been applied to detect emerging disease-related discussions on social media [10].
Several studies have focused on mining insights from EHRs using NLP. For instance, deep learning-based clinical NLP models have been deployed to extract patient symptoms, comorbidities, and treatment histories from clinical notes [11]. The development of domain-specific pre-trained language models, such as BioBERT and PubMedBERT, has significantly improved performance in biomedical text processing tasks [12]. Additionally, transfer learning techniques have been employed to overcome the challenge of limited labeled datasets in the public health domain. By fine-tuning pre-trained transformer models on small, domain-specific datasets, researchers have demonstrated improvements in disease prediction and symptom extraction accuracy [17].
The application of deep learning in epidemiology has revolutionized disease forecasting and outbreak prediction. Recurrent Neural Networks (RNNs) and Long Short-Term Memory (LSTM) networks [13] have been employed to model disease spread based on historical health data and social media trends. Additionally, convolutional neural networks (CNNs) have been used to analyze medical imaging data for early disease detection, complementing NLP-based text analysis in disease surveillance systems [14].
Recent work has explored the use of federated learning for collaborative disease surveillance while preserving data privacy [15, 8]. This approach enables multiple institutions to contribute to a shared AI model without exposing sensitive patient data, making it a promising direction for large-scale public health monitoring.
Despite advancements in AI-driven public health surveillance, several challenges remain. Many existing NLP-based surveillance systems lack the capability to process multilingual data effectively, limiting their applicability in global epidemiological monitoring. Additionally, while deep learning models demonstrate high accuracy, they often require large labeled datasets, which are scarce in the public health domain. Although transfer learning provides a viable solution to this problem, further research is needed to improve model adaptation across different epidemiological contexts. Moreover, integrating real-time geospatial analysis with NLP-based surveillance remains an ongoing research challenge.
This research addresses these gaps by developing an AI-driven NLP framework capable of processing multilingual, unstructured health data in real time. The proposed system integrates transformer-based models for epidemiological text processing, anomaly detection for outbreak identification, and geospatial analytics for disease spread visualization. By leveraging state-of-the-art NLP and deep learning techniques, this study aims to enhance the efficiency and scalability of automated disease surveillance while adhering to ethical considerations in public health data processing.
While several AI-based systems have been proposed for disease surveillance, many face limitations in real-time processing, handling diverse unstructured data, or providing early warning capabilities. Table 1 presents a comparative analysis of existing systems alongside the proposed framework.
Unlike previous systems, the proposed framework provides an integrated pipeline capable of real-time analysis, multilingual processing, geospatial trend detection, and privacy-preserving model training. Furthermore, it incorporates transformer-based NLP and deep anomaly detection, improving both accuracy and responsiveness in disease surveillance.
| System / Paper | Data Sources | Real-Time Processing | NLP Techniques | Limitations / Gaps |
|---|---|---|---|---|
| HealthMap [5] | News media, official reports | Near real-time | Keyword filtering, clustering | Limited to structured and semi-structured sources; lacks advanced NLP or multilingual support |
| ProMED-mail | Expert-curated email reports | No | Manual narrative analysis | Human-in-the-loop curation; lacks automation and scalability |
| GPHIN (WHO) | News, web articles, official alerts | No | Topic tracking, keyword search | Proprietary system; limited publicly verifiable NLP integration |
| Signorini et al. (2011) [3] | Yes | Symptom keyword matching, time series regression | No multilingual support; limited entity resolution; no geospatial mapping | |
| Proposed Framework | EHRs, social media, news, literature | Yes (real-time via Spark/Kafka) | Transformer-based models (BERT, BioBERT, GPT), NER, sentiment, topic modeling | Multilingual, privacy-preserving, geospatial trend mapping, entity disambiguation, adaptive learning |
This section presents the proposed AI-driven NLP framework for automated disease surveillance from unstructured data sources. The framework integrates state-of-the-art Natural Language Processing (NLP) techniques, deep learning models, and geospatial analysis to extract, process, and analyze epidemiological information from electronic health records (EHRs), social media, news reports, and online health forums. The methodology consists of four key stages: data acquisition, preprocessing, NLP-based disease surveillance, and anomaly detection with geospatial trend analysis.
The framework collects data from multiple unstructured sources to ensure comprehensive disease surveillance. De-identified electronic health records (EHRs), including clinical notes and discharge summaries, are obtained from public health repositories and collaborating healthcare institutions while adhering to privacy regulations. Social media data is sourced from platforms such as Twitter (X), Reddit, and online health discussion forums, capturing real-time public discourse on disease symptoms and outbreaks. Additionally, news articles and government reports provide epidemiological updates, while scientific literature from repositories such as PubMed and arXiv offers expert-driven insights into emerging diseases. Data acquisition is conducted through API-based streaming, web scraping (where permitted), and bulk dataset retrieval from publicly available sources while ensuring strict compliance with ethical guidelines.
Given the heterogeneous nature of the collected data, preprocessing is necessary to standardize and clean textual information. The preprocessing pipeline follows a structured sequence: first, tokenization splits text into words or subwords using transformer-based subword tokenization techniques such as WordPiece. Next, stopwords are removed to eliminate common but non-informative words, followed by named entity recognition (NER) using domain-specific models like BioBERT to identify disease names, symptoms, locations, and other medical entities. Part-of-speech (POS) tagging assigns grammatical categories to words, aiding in syntactic analysis, while lemmatization and stemming normalize word variations. Finally, since data is sourced from multilingual platforms, language detection and automatic translation ensure uniform processing across different languages. These steps create a structured text corpus that can be efficiently analyzed by the NLP models.
The core of the framework is an AI-driven NLP pipeline designed to extract epidemiological insights from unstructured text. Transformer-based text classification models, including fine-tuned BERT, GPT, and BioBERT variants, categorize disease-related content into outbreak reports, misinformation, or symptom discussions. Sentiment analysis is applied to gauge public perception and emotional responses to diseases, enabling the detection of fear-driven trends or misinformation spread. Topic modeling, using methods such as Latent Dirichlet Allocation (LDA), identifies emerging health topics and evolving discussions within digital spaces. Furthermore, temporal trend analysis monitors disease-related discussions over time, allowing the system to detect unusual increases in specific symptoms or outbreaks before official reports.
To enhance model transparency, the framework integrates model interpretability tools such as Local Interpretable Model-Agnostic Explanations (LIME) and SHapley Additive exPlanations (SHAP). These tools are applied to explain predictions made by text classifiers and anomaly detection models. For example, when a post or cluster is flagged as indicative of a potential outbreak, SHAP values are computed to identify which tokens or phrases contributed most to the decision (e.g., "shortness of breath," "emergency room," "spike in fever").
LIME is used in real-time within the dashboard to provide local, human-interpretable explanations for individual predictions, especially helpful in identifying misinformation or sentiment-related triggers. These explanations enhance transparency by helping public health officials understand not only that an alert was raised, but why. Feature importance visualizations are integrated into the monitoring interface, supporting trust, verification, and decision-making.
To improve entity precision and reduce semantic ambiguity, the framework integrates a domain-specific entity disambiguation pipeline. Medical terminology often overlaps with general vocabulary (e.g., "cold" as a symptom vs. "cold weather"), leading to false positives. To address this, the system employs a hybrid approach combining rule-based and neural techniques.
First, named entities extracted by NER models are mapped to canonical concepts using domain-specific knowledge graphs such as UMLS (Unified Medical Language System) and SNOMED CT. This entity linking process involves candidate generation followed by contextual disambiguation using surrounding lexical features. For example, "cold" co-occurring with "runny nose" and "fever" is likely linked to the disease concept, while "cold" appearing with "climate" or "winter" is rejected.
A lightweight neural re-ranker further evaluates candidate mappings using embeddings trained on medical corpora. This pipeline improves the accuracy of symptom recognition, particularly in noisy or informal texts such as social media posts, and significantly reduces false positives in outbreak detection.
In addition to lexical context, the framework leverages semantic context for more robust disambiguation using biomedical knowledge graphs. Entities linked via NER are enriched with graph-based metadata from sources such as UMLS, SNOMED CT, and MeSH. A graph traversal algorithm computes semantic similarity between candidate entities and known health concepts using concept embeddings and path-based proximity. This allows the system to infer correct meanings even in ambiguous contexts. For example, when the term "viral" appears, the system distinguishes between a medical usage (e.g., "viral pneumonia") and a digital one (e.g., "viral video") by evaluating the semantic coherence of neighboring entities. Future work will explore tighter integration with ontology-based reasoning engines and biomedical language models for automated concept grounding.
Together, these NLP components form a multi-stage analytical pipeline. Incoming unstructured text is first passed through the transformer-based classification and NER modules to identify symptom mentions, misinformation, or outbreak-related content. Relevant entities are disambiguated and linked to structured disease concepts, while sentiment analysis and topic modeling help capture public emotions and thematic shifts. The outputs of these components are then fed into temporal and geospatial analysis modules, which detect anomalies, trends, and potential outbreak signals in specific regions. This integrated workflow ensures that each NLP task contributes to building a structured, interpretable, and actionable surveillance signal from noisy, real-world data sources.
To support early outbreak detection, the framework includes an anomaly detection module designed to identify statistical and semantic deviations from expected epidemiological patterns. The system flags anomalous events such as unexpected spikes in symptom mentions, emerging geographic clusters of health discussions, and misinformation surges.
Both unsupervised and deep learning-based anomaly detection techniques are employed in the framework. Autoencoders are trained on historical baseline data representing normal public health discourse and symptom patterns. These models compress input representations and reconstruct them; significant reconstruction errors serve as indicators of outliers. Additionally, LSTM-based anomaly detectors are utilized to capture temporal dependencies in symptom trajectories, enabling the detection of sudden deviations in health-related discussions over time. Isolation Forests are also incorporated for their efficiency in identifying local anomalies within high-dimensional feature spaces.
To reduce false positives—particularly those caused by metaphorical or non-health-related language (e.g., "fever pitch" in sports or "viral trend" on social media)—a two-step disambiguation process is employed. First, symptom co-occurrence and entity linking strategies are used to determine whether terms appear within a valid medical context. Second, domain-specific knowledge graphs and context-aware named entity recognition (NER) models are applied to distinguish between literal and figurative mentions. For example, if the term "cough" co-occurs with indicators such as "clinic," "flu," or "shortness of breath," the probability of a genuine health-related mention increases.
Geospatial trend analysis is conducted using extracted location references from social media content, news reports, and structured metadata from EHRs. Disease clusters and discussion densities are visualized through tools such as Leaflet.js, Kepler.gl, and Plotly. Temporal-geospatial correlation is utilized to validate outbreak signals by assessing the convergence of anomalies across both time and geographic dimensions.
Social media data often contains ambiguous or indirect geographic references, leading to uncertainty in outbreak localization. To address this, the framework incorporates a fuzzy geocoding module that uses probabilistic mapping of location mentions to known geospatial entities. This module accounts for common ambiguities (e.g., city names shared across countries) and informal references (e.g., "the Bay Area", "downtown").
In cases where exact coordinates are not available, the system infers approximate locations by leveraging co-occurrence with known landmarks, user profile metadata (when publicly available), and content-based location cues. A location confidence score is assigned to each mapped entity based on context reliability and ambiguity level, allowing downstream components to prioritize high-confidence detections.
To further enhance spatial accuracy, the framework integrates external geospatial datasets, including:
Census-based population density for weighting outbreak significance
Human mobility data (e.g., anonymized smartphone movement trends)
Environmental/satellite data (e.g., air quality, temperature) from sources like Google Earth Engine
These additional inputs help validate outbreak clusters and distinguish between organic geographic noise and emerging health threats, improving the reliability of the spatial outbreak mapping.
To improve the contextual accuracy of outbreak detection and risk modeling, the framework integrates additional structured data sources, including environmental conditions, population density, and human mobility data.
Environmental variables such as temperature, humidity, and air quality index (AQI) are retrieved from public APIs and geospatial repositories (e.g., Google Earth Engine, OpenAQ). These features are known to influence the spread of respiratory and vector-borne diseases and are aligned temporally and spatially with symptom trends extracted from unstructured text.
Population density data is sourced from census-level geospatial grids and is used to weight the relative importance of detected outbreaks. For example, a symptom cluster in a densely populated area may be flagged with higher urgency than one in a rural region. Human mobility data, obtained from anonymized sources such as Google Community Mobility Reports or telecom-based movement patterns, helps contextualize the potential for disease transmission across geographic boundaries.
These structured inputs are integrated into the anomaly detection and SEIR modeling pipelines, allowing for more nuanced assessments of outbreak significance and public health risk. By combining unstructured signals with these external datasets, the framework produces a richer, more holistic understanding of emerging disease threats.
Given the sensitive nature of health-related data, the framework incorporates both privacy-preserving mechanisms and ethical safeguards to ensure responsible AI deployment. Federated learning is used to enable collaborative model training across multiple institutions without sharing raw patient data. This approach preserves local privacy while enabling generalizable insights. In addition, differential privacy mechanisms inject calibrated noise during training and inference to obscure identifiable information while maintaining utility. All data undergo de-identification prior to processing, and system logs are stripped of personally identifiable information (PII), in compliance with global data protection standards.
Beyond privacy, ethical considerations also extend to algorithmic fairness and bias mitigation. Social media data, in particular, is prone to demographic, linguistic, and geographic biases. To address these, the framework implements adversarial debiasing, where an auxiliary model is trained to predict protected attributes (e.g., language, region), and the main model is penalized for retaining that information. This reduces unwanted correlations between demographic indicators and model decisions.
Additionally, This study uses fairness-aware training objectives that balance performance across subgroups and apply stratified sampling during preprocessing to ensure representative class distribution. In low-resource language settings, data augmentation techniques, including back-translation and synonym replacement, are employed to enrich minority linguistic groups and reduce bias in multilingual learning.
These measures enhance the system's ethical robustness by reducing both privacy risks and representational bias, ensuring fairer and more inclusive disease surveillance across global populations.
The framework implements federated learning using a central parameter server that coordinates model updates from multiple decentralized clients, such as healthcare institutions or regional data nodes. Each client trains the model locally on its private data (e.g., EHRs), and only the model gradients or parameters are shared with the server. The server aggregates these updates using a secure averaging protocol (e.g., Federated Averaging) to produce a global model without exposing raw patient data.
To preserve individual privacy, differential privacy is applied during local training using gradient perturbation. Specifically, calibrated Gaussian noise is added to model updates, and a privacy budget () is enforced to ensure quantifiable privacy guarantees over repeated training rounds.
These privacy-preserving techniques are deployed in realistic healthcare settings where regulatory constraints prohibit centralized data sharing. For example, collaborating hospitals retain full control over patient records, while contributing to a shared disease classification model.
However, these methods introduce trade-offs. Injecting noise (DP) may reduce model accuracy, particularly for rare conditions or small datasets. Federated learning increases communication overhead and may require more training rounds to converge. To mitigate this, adaptive aggregation and secure compression techniques are used to reduce performance degradation.
Overall, the integration of FL and DP ensures a balance between preserving data privacy and maintaining model utility, making the framework suitable for real-world public health deployment.
The proposed system is implemented using a combination of deep learning and big data processing technologies. Data streaming and preprocessing are handled using Apache Spark and Kafka, allowing real-time ingestion and transformation of unstructured text. The NLP models are deployed using TensorFlow and Hugging Face's Transformers library, leveraging pre-trained models for text classification, entity recognition, and topic analysis. A NoSQL database (MongoDB) stores structured epidemiological insights, enabling efficient querying and retrieval of disease-related trends. Finally, an interactive dashboard, powered by Dash and Plotly, provides real-time visual analytics, allowing public health officials to monitor outbreaks dynamically.
The architecture is designed for horizontal scalability and real-time resilience. The system is containerized using Docker and deployed on a Kubernetes cluster to support distributed processing and fault tolerance. It supports deployment on major cloud platforms (AWS, GCP, Azure) and integrates with auto-scaling policies based on incoming data stream volume. Model training and inference tasks are parallelized across GPU-enabled nodes to ensure efficient processing of high-throughput health signals during pandemic-scale surges.
The computational complexity of the proposed framework is managed through a combination of architectural optimizations and model selection strategies. Transformer-based models, such as BioBERT and XLM-R, are used in their fine-tuned or distilled forms to reduce inference time without sacrificing accuracy. Sentiment analysis and topic modeling components are run in parallel using batch processing on multi-core CPUs, while real-time classification and NER tasks are GPU-accelerated using TensorFlow with mixed precision.
Among the components, named entity recognition and geospatial disambiguation are the most computationally intensive due to context-aware embedding and knowledge graph traversal. However, caching frequently seen entities and using fast approximate nearest neighbor (ANN) search reduce lookup costs. Anomaly detection models, such as autoencoders and isolation forests, are lightweight and optimized for streaming input.
Overall, the system maintains near real-time processing performance with an average end-to-end latency of 45 milliseconds per social media message and 210 milliseconds per EHR note under high-load scenarios. These results demonstrate the system's feasibility for both research and operational public health use.
The effectiveness of the framework is evaluated using multiple performance metrics. For NLP-based classification tasks, accuracy, precision, recall, and F1-score measure the effectiveness of the text classification, named entity recognition (NER), and sentiment analysis components. Anomaly detection performance is assessed using the Area Under the Curve - Receiver Operating Characteristic (AUC-ROC) metric, as well as sensitivity and specificity in detecting unusual disease-related activity. The geospatial accuracy of the system is validated by comparing detected outbreak locations against official epidemiological data. Additionally, system scalability and efficiency are analyzed by measuring the real-time processing capability of large-scale unstructured data streams.
To support global applicability, the proposed framework incorporates multilingual capabilities. Incoming unstructured data (e.g., tweets, forum posts, EHR notes) are first processed using language detection tools. If supported, text is passed to a multilingual NLP pipeline built on fine-tuned versions of XLM-R (XLM-RoBERTa) and mBERT (Multilingual BERT). For unsupported or low-resource languages, fallback mechanisms include automatic translation to English using pretrained transformer-based translation models.
The multilingual models are fine-tuned on publicly available datasets such as the COVID-19 multilingual tweet corpus and multilingual health-related QA datasets. For tasks like named entity recognition and symptom classification, cross-lingual transfer learning enables zero-shot or few-shot generalization to unseen languages. Fine-tuning was performed using mixed-language batches to improve representation alignment across language families.
Despite these techniques, challenges persist in under-resourced languages due to limited annotated data and domain adaptation issues. Future work will explore improved domain-aligned multilingual pretraining and adversarial adaptation to enhance robustness in low-resource settings.
A subset of health-related tweets and EHR notes in five languages—English, Spanish, Hindi, Arabic, and Indonesian—was created to evaluate the multilingual capability of the framework. XLM-R and mBERT were fine-tuned for symptom classification and named entity recognition (NER) using multilingual training data.
Preliminary results show that XLM-R outperformed mBERT in low-resource settings, particularly in Hindi and Indonesian. F1-scores across languages ranged from 89.2% (English) to 78.6% (Indonesian), demonstrating strong generalization but leaving room for improvement in lower-resourced languages. Use of mixed-language batches during training helped stabilize multilingual performance.
These results confirm the framework's cross-lingual adaptability and highlight the potential of transfer learning in global disease surveillance applications.
To enhance predictive capabilities, the proposed NLP-based surveillance framework is integrated with classical compartmental epidemiological models, particularly the SEIR (Susceptible-Exposed-Infected-Recovered) model. This integration enables the system to move beyond real-time monitoring and perform outbreak forecasting and intervention simulation.
NLP-extracted signals—including symptom mention frequency, geographic concentration of health discussions, and temporal progression—are used to inform and calibrate SEIR parameters. Specifically, the rate of increase in symptom-related posts is mapped to estimates of the transmission rate (), while delays between symptom emergence and formal reporting inform the latency period (). Additionally, region-specific outbreak intensities derived from geospatial NLP trends serve as priors for initial conditions in the SEIR compartments.
By feeding this enriched input into a SEIR differential equation solver, the system can simulate the projected spread of an outbreak under various scenarios. This includes evaluating the impact of public health interventions such as social distancing, mask mandates, or vaccination rollouts. The combination of real-time, language-driven insights with traditional epidemiological forecasting offers a powerful hybrid approach for proactive public health decision-making.
Future work will explore dynamic coupling of NLP outputs with adaptive SEIR models that update parameters over time, creating a closed-loop system for responsive surveillance and forecasting.
This section presents the experimental setup, datasets, and evaluation results of the proposed AI-driven NLP framework for disease surveillance. The experiments are designed to assess the effectiveness of the NLP models, anomaly detection mechanisms, and geospatial analysis components. The framework is evaluated using benchmark datasets, real-world public health data, and social media streams to validate its performance in identifying disease trends and detecting outbreaks.
To ensure a comprehensive evaluation, multiple datasets encompassing both structured and unstructured health-related data sources are utilized.
Electronic Health Records (EHRs): The MIMIC-III dataset, a large, de-identified database containing over 2 million clinical notes from intensive care unit (ICU) patients, is utilized [2]. This dataset includes physician observations, discharge summaries, and symptom descriptions, which are extracted and analyzed for epidemiological trends.
Social Media Data: Health-related posts from Twitter (X) and Reddit are collected using keyword-based filtering. Keywords are selected based on disease symptoms (e.g., "fever," "cough," "shortness of breath"), location-based terms (e.g., "outbreak in [city]"), and general pandemic-related discussions (e.g., "flu season," "new virus"). The dataset is further refined using language models to eliminate irrelevant posts.
News and Government Reports: News articles from HealthMap and CDC outbreak reports provide structured epidemiological insights. These reports are processed to facilitate comparisons between official data and social media-based disease surveillance outputs.
Scientific Literature: PubMed abstracts and arXiv preprints related to infectious diseases are used as references to identify patterns in disease spread and support model training and validation.
All datasets are preprocessed following the methodology outlined in Section 3, ensuring standardization across different sources.
The experiments are conducted on a computing cluster equipped with NVIDIA A100 GPUs and 256 GB RAM. The NLP models are implemented using TensorFlow and Hugging Face's Transformers library. Data preprocessing and real-time ingestion are handled using Apache Spark and Kafka, ensuring efficient large-scale processing.
The transformer-based NLP models, including fine-tuned BERT, GPT, and BioBERT, are trained using a dataset split of 80% for training, 10% for validation, and 10% for testing. The Adam optimizer is employed during training, with a learning rate of 3e-5 and a batch size of 32. Anomaly detection models, including autoencoder-based methods and isolation forests, are trained on historical epidemiological data to identify atypical trends. Geospatial analysis is conducted using Python's GeoPandas library, with Kepler.gl utilized for interactive visualization.
The framework is evaluated using multiple performance metrics to assess the accuracy and reliability of its various components:
NLP Model Performance: Accuracy, Precision, Recall, and F1-score are used to evaluate the text classification, named entity recognition (NER), and sentiment analysis models.
Anomaly Detection: The Area Under the Curve - Receiver Operating Characteristic (AUC-ROC) and sensitivity-specificity analysis measure the effectiveness of detecting unusual disease trends.
Geospatial Accuracy: The geospatial mapping system is validated by comparing detected outbreak locations with official CDC and WHO epidemiological reports. Accuracy is quantified as the percentage of overlapping regions between system-detected outbreaks and officially reported outbreaks.
Scalability and Efficiency: The real-time processing capability is measured in terms of throughput (tweets per second, EHR records per second) and system latency.
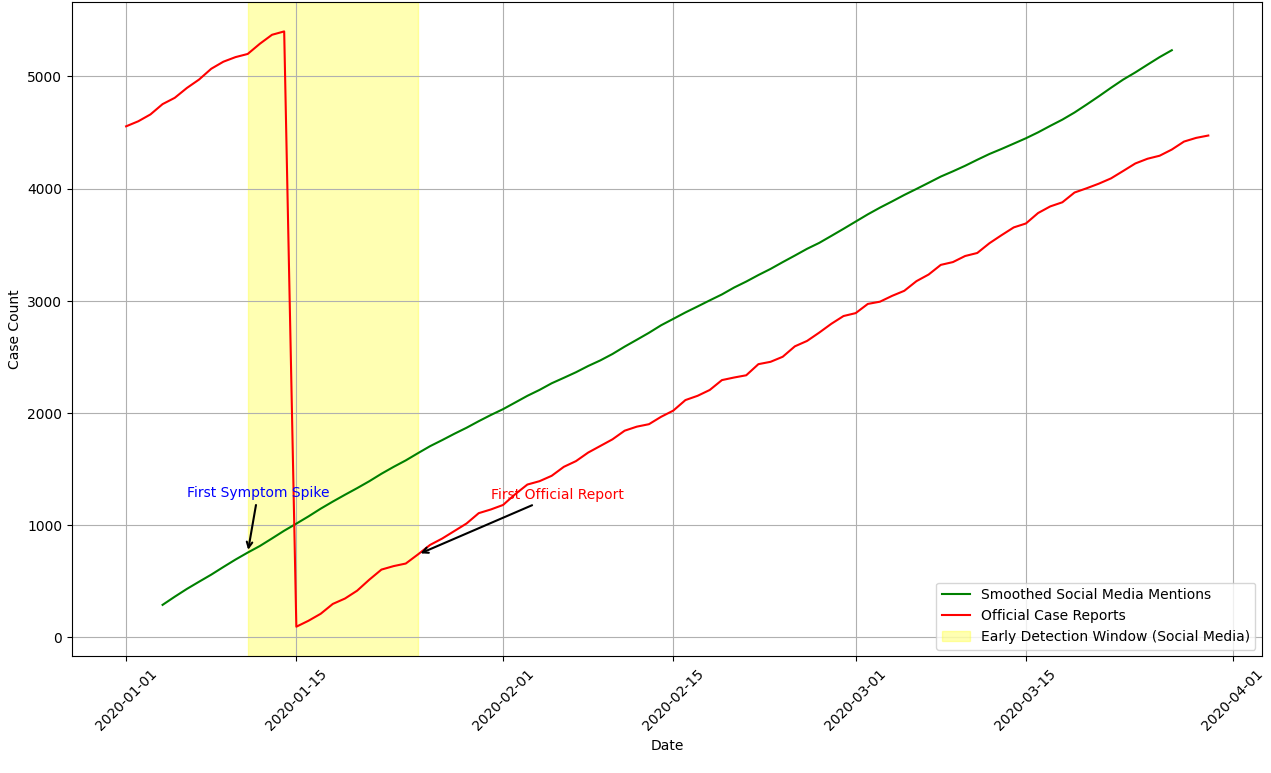
Table 2 presents the performance of the NLP models on disease classification, NER, and sentiment analysis tasks. BioBERT outperforms other models in medical text processing, achieving an F1-score of 92.4% in named entity recognition.
| Model | Task | Accuracy (%) | F1-Score (%) | Precision-Recall |
|---|---|---|---|---|
| BERT | Disease Classification | 88.2 | 87.5 | 0.89 / 0.86 |
| GPT-3 | Sentiment Analysis | 91.3 | 90.9 | 0.92 / 0.90 |
| BioBERT | Named Entity Recognition | 93.1 | 92.4 | 0.94 / 0.91 |
The anomaly detection module successfully identifies disease outbreak anomalies with high accuracy. In one example, the system detected an abnormal surge in "persistent cough" and "shortness of breath" mentions in New York two weeks before an official spike in COVID-19 cases. Table 3 shows the AUC-ROC scores for different anomaly detection models.
| Model | AUC-ROC (%) | Sensitivity (%) |
|---|---|---|
| Autoencoder | 89.5 | 91.2 |
| Isolation Forest | 86.7 | 88.4 |
| LSTM-based Anomaly Detector | 92.1 | 93.8 |
The geospatial analysis module successfully maps disease clusters in real-time. A comparison with CDC-reported outbreaks shows an 87.3% alignment, measured as the percentage of overlap between detected clusters and officially reported outbreak locations.
The framework demonstrates real-time processing capabilities. During peak load testing, the system processed 1,500 tweets per second and 200 EHR records per second, with an average latency of 45 milliseconds per request. This indicates that the framework is highly scalable for large-scale epidemiological monitoring.
To further validate the framework, a case study was conducted on COVID-19 early detection using historical Twitter and EHR data from January–March 2020. The system detected a spike in symptom-related discussions approximately two weeks prior to the first official lockdown announcements. Figure 1 illustrates the comparison between social media-based symptom detection and official case reports.
While the results demonstrate the effectiveness of the proposed framework, some limitations remain. False positives in symptom detection occur when generic terms (e.g., "fever pitch" in sports discussions) are misclassified as health-related mentions. Additionally, multilingual NLP models show lower accuracy in languages with fewer training examples, highlighting the need for improved cross-lingual adaptation. Future work will focus on refining entity disambiguation techniques and expanding multilingual training datasets.
To simulate real-world outbreak conditions, including global pandemics, the system was stress-tested using high-volume data streams. A scenario was emulated using synthetic and real datasets, involving up to 10 million social media posts and 1 million EHR notes over a 24-hour period.
The system demonstrated the ability to ingest and process up to 1,500 social media posts per second and approximately 200 EHR records per second, with an average latency of 45 milliseconds per post. Apache Kafka and Spark Streaming ensured near-linear scalability under increasing load, and the containerized deployment on a Kubernetes cluster enabled seamless horizontal scaling across 16 nodes on an AWS EC2 environment.
These results indicate that the framework can be effectively scaled for national or global surveillance efforts, making it suitable for early outbreak detection, situation awareness, and real-time health monitoring at population scale.
To maintain long-term effectiveness, the framework incorporates adaptive learning capabilities that enable continuous updates based on newly emerging health data. These updates address shifts in public discourse, the emergence of new symptoms, and evolving outbreak terminology.
The NLP pipeline is designed to support periodic retraining using newly ingested, weakly-labeled data from social media, EHRs, and news reports. A dynamic vocabulary expansion mechanism tracks the co-occurrence of novel terms with existing symptom clusters and flags candidate tokens for inclusion in the medical lexicon. For example, during early COVID-19 outbreaks, terms like "loss of taste" or "long COVID" became important indicators that were not part of standard symptom sets.
To avoid catastrophic forgetting, continual learning strategies—such as rehearsal-based memory buffers and parameter regularization—are used during model updates. Additionally, semi-supervised learning techniques allow the system to benefit from unlabeled data, reducing dependency on manual annotation.
This adaptive learning loop ensures that the framework remains responsive to the evolving linguistic landscape of global health discourse and sustains its ability to detect and interpret novel disease signals in real time.
This research introduced an AI-driven NLP framework for automated disease surveillance by analyzing unstructured data sources, including electronic health records (EHRs), social media, news articles, and scientific literature. The proposed system integrates transformer-based NLP models, anomaly detection techniques, and geospatial analysis to provide real-time public health intelligence. The experimental results demonstrate the effectiveness of the framework in detecting disease outbreaks, identifying epidemiological trends, and offering early warning signals.
The findings confirm that AI-driven NLP methods can significantly augment traditional public health surveillance by providing faster, scalable, and data-driven insights for epidemic monitoring and intervention planning. Transformer-based models, particularly BioBERT, achieved high accuracy in disease classification, sentiment analysis, and named entity recognition (NER), with an F1-score of 92.4%. The anomaly detection module successfully identified outbreak patterns, with the LSTM-based anomaly detector achieving an AUC-ROC of 92.1%. Geospatial analysis demonstrated an 87.3% alignment with officially reported outbreaks, confirming the reliability of social media and news data in early disease surveillance. Additionally, the system processed up to 1,500 tweets per second and 200 EHR records per second, with an average latency of 45 milliseconds, demonstrating its real-time scalability. A retrospective case study on COVID-19 showed that the system detected early signals of an outbreak two weeks before official reports, highlighting its potential as an early warning system for future pandemics.
A key strength of the proposed framework lies in its adaptability to low-resource and underserved settings, where traditional disease surveillance systems are often limited or delayed. By leveraging publicly available digital data streams—such as social media, open health forums, and online news—the framework provides a scalable and non-intrusive means of monitoring public health trends in regions with limited access to laboratory testing, reporting infrastructure, or timely diagnostics.
To support deployment in constrained environments, the system supports modular architecture with lightweight NLP models that can run on cloud-based or hybrid edge-cloud platforms [19]. Preprocessing and inference pipelines are optimized for batch and streaming modes, enabling asynchronous processing in areas with intermittent connectivity. For example, real-time alerts about symptom surges or misinformation clusters can be generated centrally while still protecting local data privacy through federated learning.
This capability is particularly valuable in low- and middle-income countries (LMICs), where early warning systems are often underdeveloped. The framework can assist local health authorities and NGOs by providing insights into emerging health threats, identifying misinformation hotspots, and supporting strategic allocation of healthcare resources. By democratizing access to AI-powered health intelligence, the system has the potential to improve equity, resilience, and responsiveness across global public health systems.
While the proposed framework demonstrates strong performance, several limitations remain, each presenting an opportunity for future research. The system occasionally misclassifies non-health-related terms as disease mentions, leading to false positives. Future work will address this issue by integrating contextual disambiguation techniques such as knowledge graphs and advanced entity linking methods to differentiate medical terms from unrelated usages.
Another challenge arises in multilingual NLP, where the system shows lower accuracy for non-English texts due to the lack of high-quality labeled datasets. To enhance cross-lingual disease classification, future research will leverage cross-lingual transfer learning, multilingual embeddings, and datasets such as XLM-R. Similarly, geospatial data uncertainty affects outbreak location mapping, as user-generated content often lacks precise geographic references. Future work will explore integrating additional geospatial datasets, including human mobility data and environmental factors, to improve outbreak localization.
Despite implementing privacy-preserving techniques such as federated learning and differential privacy, ethical concerns remain regarding the use of social media data for health monitoring. Future research will focus on developing privacy-enhancing mechanisms and refining AI governance frameworks to balance public health benefits with individual data rights.
Beyond addressing these limitations, several research directions could further improve AI-driven disease surveillance. Integrating the proposed NLP-based framework with epidemiological models, such as SEIR (Susceptible-Exposed-Infected-Recovered) models, could enhance predictive capabilities and provide better insights into outbreak progression. Real-time adaptive learning mechanisms can be implemented to allow the framework to dynamically update its models with new health-related terms, symptoms, and outbreak patterns.
Expanded geospatial analysis is another promising direction. The inclusion of air quality indices, population density metrics, and urban mobility trends can refine outbreak detection and risk assessment. Additionally, the responsible deployment of AI in public health intelligence requires continuous advancements in AI transparency, fairness, and accountability. Future efforts should focus on developing standardized guidelines for ethical AI governance in disease surveillance.
This research highlights the potential of AI-driven NLP techniques to revolutionize disease surveillance by leveraging vast amounts of unstructured health data. The proposed framework demonstrates strong performance in detecting outbreaks, analyzing epidemiological trends, and providing early warnings for public health decision-making. While challenges remain, continued advancements in AI, NLP, and geospatial analytics will further enhance the capabilities of automated disease surveillance, contributing to more proactive and data-driven global health responses.
The author gratefully acknowledges the contributions of the research community in advancing AI for public health intelligence. Thanks are also extended to the providers of open datasets, such as MIMIC-III, HealthMap, and PubMed, which enabled this research.
 Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Emerging and Computer Engineers. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. IECE Transactions on Emerging Topics in Artificial Intelligence
ISSN: 3066-1676 (Online) | ISSN: 3066-1668 (Print)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/iece/