Abstract
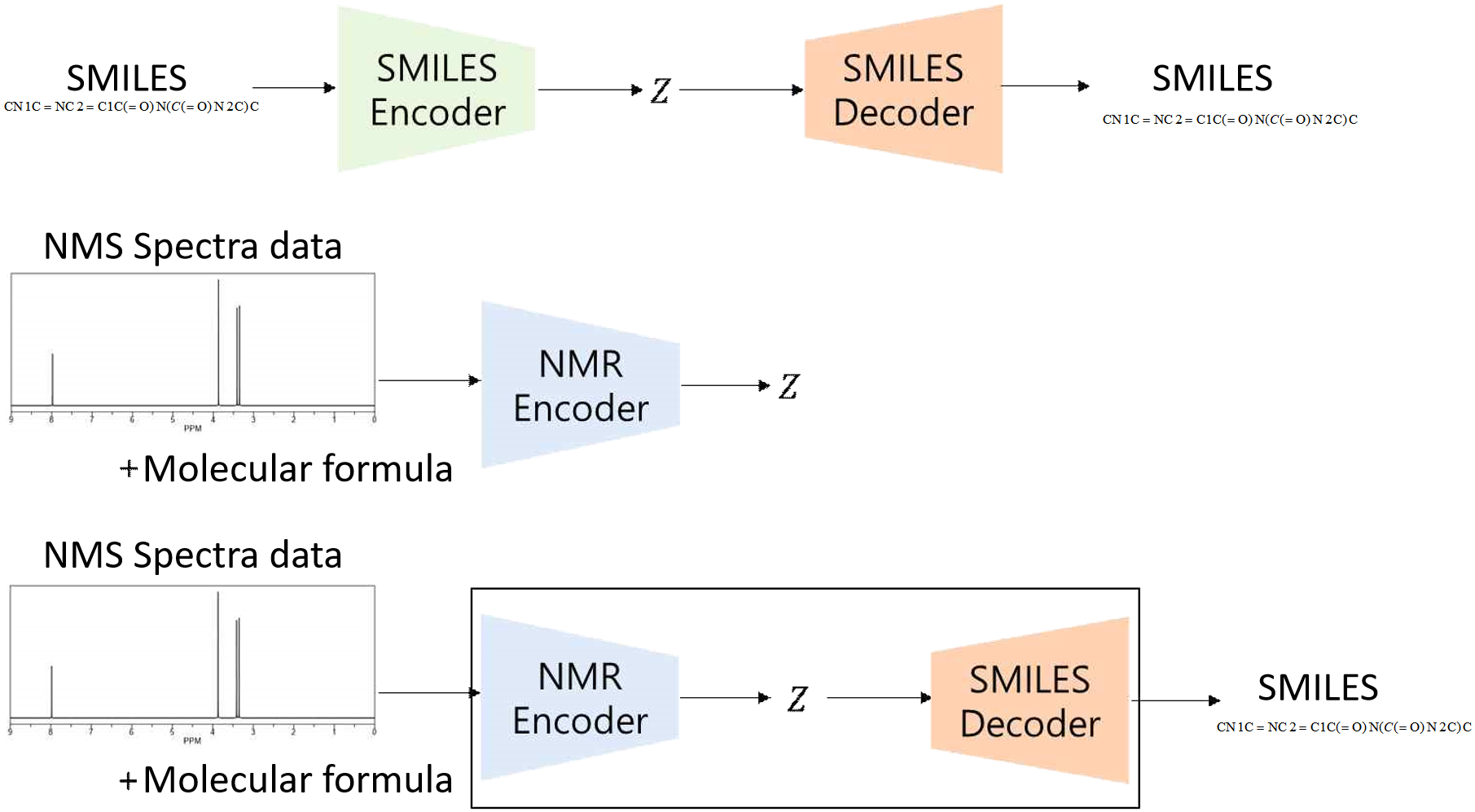
Interpreting NMR spectra to accurately predict molecular structures remains a significant challenge in chemistry due to the complexity of spectral data and the need for precise structural elucidation. This study introduces NMRGen, a generative modeling framework that predicts molecular structures from NMR spectra and molecular formulas. The framework combines a SMILES autoencoder (GRU-based encoder-decoder) and an NMR encoder (CNN and DNN layers) to map spectral data to molecular representations. The SMILES autoencoder compresses and reconstructs SMILES strings, while the NMR encoder processes NMR spectra to generate latent vectors aligned with those from the SMILES encoder. Experiments were conducted using NMR spectra and SMILES datasets. The model was trained in three stages: (1) training the SMILES autoencoder, (2) aligning latent vectors from the NMR encoder, and (3) simultaneous training of both components. Results revealed that while the SMILES autoencoder performed adequately, the NMR encoder struggled to map spectral data effectively. Most generated SMILES strings were invalid, with valid ones primarily consisting of carbon chains (e.g., CCC...C). The Tanimoto coefficient between generated and target molecules ranged from 0.1 to 0.2, indicating low similarity. Despite these limitations, NMRGen demonstrates the potential of generative models for molecular structure prediction. Future work will focus on improving performance through larger datasets, advanced loss functions, and enhanced architectures.
Funding
This work was supported without any funding.
Cite This Article
APA Style
Vavekanand, R. (2025). NMRGen: A Generative Modeling Framework for Molecular Structure Prediction from NMR Spectra. IECE Transactions on Emerging Topics in Artificial Intelligence, 2(1), 16–25. https://doi.org/10.62762/TETAI.2024.277656
Publisher's Note
IECE stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Institute of Emerging and Computer Engineers (IECE) or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue